Key Points
CD62Ldim neutrophils seen in acute inflammation are of an age similar to that of segmented neutrophils; banded neutrophils are immature cells.
CD62Ldim neutrophils cluster separately from banded and normal segmented neutrophils based on proteome profiling.
Abstract
During acute inflammation, 3 neutrophil subsets are found in the blood: neutrophils with a conventional segmented nucleus, neutrophils with a banded nucleus, and T-cell–suppressing CD62Ldim neutrophils with a high number of nuclear lobes. In this study, we compared the in vivo kinetics and proteomes of banded, mature, and hypersegmented neutrophils to determine whether these cell types represent truly different neutrophil subsets or reflect changes induced by lipopolysaccharide (LPS) activation. Using in vivo pulse-chase labeling of neutrophil DNA with 6,6-2H2-glucose, we found that 2H-labeled banded neutrophils appeared much earlier in blood than labeled CD62Ldim and segmented neutrophils, which shared similar label kinetics. Comparison of the proteomes by cluster analysis revealed that CD62Ldim neutrophils were clearly separate from conventional segmented neutrophils despite having similar kinetics in peripheral blood. Interestingly, the conventional segmented cells were more related at a proteome level to banded cells despite a 2-day difference in maturation time. The differences between CD62Ldim and mature neutrophils are unlikely to have been a direct result of LPS-induced activation, because of the extremely low transcriptional capacity of CD62Ldim neutrophils and the fact that neutrophils do not directly respond to the low dose of LPS used in the study (2 ng/kg body weight). Therefore, we propose CD62Ldim neutrophils are a truly separate neutrophil subset that is recruited to the bloodstream in response to acute inflammation. This trial was registered at www.clinicaltrials.gov as #NCT01766414.
Introduction
Neutrophils engage in the resolution of infection by phagocytosing and killing pathogens. Decreased functionality of neutrophils can lead to infectious complications and even sepsis.1 Conversely, hyperactivation can lead to severe proinflammatory conditions such as acute respiratory distress syndrome. Neutrophils are involved in the balance of hyper- and hypoactivation of the immune system by releasing proinflammatory and toxic mediators2,3 and by suppressing excessive immune responses.4
Acute inflammation induces recruitment of neutrophil phenotypes into the peripheral blood that are not present in either circulation or the marginated pool during homeostasis.5 Several studies have identified such inflammatory phenotypes focusing on different characteristics, such as differential expression of 3-fucosyl-N-acetyl-lactosamine (CD15) and Fc-γ receptor-III (CD16),6 interleukin-4Rα (CD124) expression,7 low buoyant density,8 arginase expression,9 or immunomodulatory functionality.10 On the basis of the differential expression of CD16 and L-selectin (CD62L),11 we have identified at least 3 different subsets of inflammatory neutrophils: (1) neutrophils with a segmented nuclear morphology resembling prototypically mature blood neutrophils characterized by CD16bright/CD62Lbright expression (segmented), (2) CD16dim/CD62Lbright cells with a banded nuclear morphology (banded), and (3) CD16bright/CD62Ldim cells with on average a larger number of nuclear lobes (although not as high as in foliate deficiency–induced hypersegmentation) that are capable of suppressing T-cell proliferation (CD62Ldim).11
The 3 neutrophil subsets have been observed in a range of acute inflammatory diseases, including sepsis,11 severe trauma,11 burns, and cancer.12 Because of their opposing pro- and anti-inflammatory functions, manipulation of neutrophil subset release and development might allow for steering the acute immune response. This can be achieved, for example, by activating CD62Ldim neutrophils during hyperinflammatory conditions or by inhibiting CD62Ldim neutrophils to counteract the compensatory anti-inflammatory response syndrome in severely ill patients.13 Very little is known about the origins of these subsets, however. It is commonly thought that the increase in nuclear segmentation is associated with increasing cellular age,4,14 because banded neutrophils have been shown to be younger cells in patients with cancer,15,16 and CD62Ldim neutrophils have been shown to be aged cells in mice.17 In humans, however, hypersegmented neutrophils that are observed in pernicious anemia are thought to directly mature from banded cells into hypersegmented cells.18 In addition, in vivo 3H-thymidine labeling in a patient with pernicious anemia showed that labeled hypersegmented neutrophils started to appear in circulation at the same time as normal neutrophils, although the peak in labeling of the hypersegmented neutrophils was observed 1 day later. Taken together, the conflicting data on CD62Ldim neutrophils in mice and hypersegmented neutrophils in patients with pernicious anemia illustrate that it remains an open question whether the CD62Ldim neutrophil subset observed in acute inflammation represents aged cells.
All 3 subsets are detected in the bone marrow during homeostasis,19,20 but little is known regarding the presence of these cells outside the bone marrow or their relative numbers at the different tissue sites. The human endotoxemia model is a valuable model to study acute systemic inflammation21 and induces changes in blood immune cells reminiscent of those found in septic22,23 or severely injured patients.24 These conditions induce the recruitment of banded and CD62Ldim neutrophils into the blood. Transcriptome analyses of neutrophil subsets in the human endotoxemia model25,26 revealed large differences between banded and CD62Ldim neutrophils, but these studies did not relate these differences to the segmented neutrophil subset. Moreover, characterization of neutrophil subsets by transcriptome analysis is hampered by the fact that these cells have a low transcriptional capacity,27 and many changes in protein expression levels may not be reflected by changes in messenger RNA (mRNA) expression. For example, most granule proteins are produced and stored during maturation in the bone marrow3,28 and can be lost by degranulation. These changes will be missed by mRNA analysis.
Our study was designed to gain more insight into the properties and origins of the 3 neutrophil subsets found in acute inflammation. In vivo pulse-chase deuterium 2H-labeling was performed to determine the maturation times of the different subsets observed during acute inflammation. In addition, we performed high-end proteomic analysis on purified blood neutrophil subsets to assess differences among the 3 subsets.
Materials and methods
Participants
Samples were obtained from 20 healthy male volunteers who participated in a human endotoxemia trial (registered at www.clinicaltrials.gov as #NCT01766414), in which 10 volunteers were treated with C1-esterase inhibitor and 10 were treated with a placebo. The study was approved by the ethics review board of the Radboud University Medical Centre and conducted in compliance with the Declaration of Helsinki (adapted by World Medical Association General Assembly, Fortaleza, Brazil, 2013), International Conference on Harmonisation Good Clinical Practice guidelines, and the rulings of the Dutch Medical Research Involving Human Subjects Act. Written informed consent was obtained from all study participants. Participants were screened and found healthy based on physical examination, electrocardiography, and hematological laboratory values. All volunteers were negative for HIV and hepatitis B antibodies in serum. Participants with febrile illness <2 weeks before LPS injection were excluded, as were those taking prescription drugs. Participants were asked to refrain from caffeine and alcohol use in the 24 hours before LPS challenge and to refrain from food intake 12 hours before LPS challenge.
In vivo 2H labeling
The relative age of neutrophil subsets was determined using in vivo 2H labeling, as described previously.29,30 In short, all 20 volunteers were asked to drink 12 doses every 30 minutes of 6.67 g of metabolic-grade 6,6-2H2-glucose (total, 80 g; Cambridge Isotope Laboratories, Tewksbury, MA) at 3 to 11 days before LPS administration (as described in the following paragraph). The 6,6-2H2-glucose label is metabolized and incorporated into the DNA of all dividing cells via the de novo nucleotide synthesis pathway.17,18 During the labeling procedure, blood was collected by finger prick to determine the availability of label in plasma, as described previously.30
Experimental endotoxemia model
LPS challenge was performed according to a strict protocol, as described previously.31 In short, after admission to the research medium care unit of the Radboud Medical Centre, heart rate and blood pressure were monitored starting at t = −1 hour until discharge at t = 8 hours. A cannula was placed in an antecubital vein to permit infusion of hydration fluid (1.5 L; 2.5% glucose, 0.45% saline) from t = −1 hour until t = 0 hours. Then a single dose of 2 ng per kilogram of bodyweight LPS (US Standard Reference Endotoxin Escherichia coli O:113, obtained from the Pharmaceutical Development Section of the National Institutes of Health, Bethesda, MD) was injected (t = 0 hours). LPS injection was followed by continuous infusion of hydration fluid (2.5% glucose, 0.45% saline; 150 ml/h) from t = 0 to t = 8 hours. The course of LPS-induced symptoms (headache, shivering, nausea, vomiting, muscle pain, and back pain) was scored every 30 minutes on a 6-point Likert scale (0 = no symptoms; 5 = very severe symptoms; vomiting = 3 points), adding up to a maximum of 28 points.32 White blood cell counts were analyzed on a routine hematocytometer at t = 0, 1, 2, 6, and 8 hours.
Neutrophil isolation
Leukocytes were isolated from blood anticoagulated with sodium heparin 5 minutes before injection of LPS and at 1, 3, 6, and 8 hours after LPS injection. After erythrocyte lysis in ice-cold isotonic erythrocyte lysis buffer (150 mM NH4Cl, 10 mM KHCO3, and 0.1 mM NA2EDTA), leukocyte preparations were stained with antibodies in phosphate-buffered saline supplemented with 0.32% trisodium citrate and 10% human pasteurized plasma solution (Sanquin, Amsterdam, The Netherlands): CD14-ECD (clone RMO52; Beckman Coulter, Pasadena, CA) and CD62L-PE (clone SK11) and CD16-Alexa647 (clone 3G8; both from BD Biosciences, San Jose, CA). Neutrophil subsets were sorted using an AriaIII fluorescence-activated cell sorter (FACS; BD Biosciences). Neutrophils were identified based on a forward scatter/side scatter granulocyte gate; doublet exclusion was based on side scatter height/width and CD14− expression. Subsequently, neutrophil subsets were sorted as CD16dim/CD62Lbright (banded), CD16bright/CD62Lbright (segmented), and CD16bright/CD62Ldim (CD62Ldim; supplemental Figure 1, available on the Blood Web site). Sorted populations were reanalyzed and typically >99% pure.
Neutrophil nuclear morphology
Neutrophil nuclear morphology was determined by manual counting of May-Grünwald-Giemsa–stained cytospins by an experienced technician in a blinded manner. The microscope used was a Leica DMRXE microscope equipped with a 100× oil immersion objective (Leica Microsystems, Wetzlar, Germany). A strict definition was used to identify a separation between lobes in such a way that these were only scored when the connection between lobes was <33% of the width of the adjacent lobes.
Determination of 2H enrichment
DNA was isolated from sorted cell populations using a NucleoSpin blood kit (Macherey-Nagel, Düren, Germany) and hydrolyzed and derivatized to pentafluoro triacetate, as described in detail previously.30 Relative quantities of unlabeled and 2H-labeled adenosine–derived pentafluoro triacetate were determined with an Agilent 7980A/5975C GC-MS in negative chemical ionization mode scanning for m/z 435 (M+0, unlabeled) and m/z 437 (M+2, labeled). Resulting enrichments were corrected for natural background enrichment and availability of 6,6-2H2-glucose in plasma, as described previously.30
Protein identification and quantification
A detailed description of the proteomic approach and western blot analyses of several candidate proteins can be found in supplemental Methods and in supplemental Figure 2. In short, 3 × 106 cells of the different neutrophil subsets were isolated from blood derived from 3 volunteers in the placebo group 3 hours after LPS infusion, because at this time point, the largest number of cells from the different subsets were present. Cells of each subset were digested with trypsin, labeled with a different label for each subset, and pooled in a 1:1:1 ratio for each individual participant. This approach allows for internally controlled quantification of the mass spectrometry data.
Samples were fractionated using strong cation exchange chromatography. Fractions were analyzed using an LTQ-Orbitrap liquid chromatography–tandem mass spectrometry (LC-MS/MS).33 Peptides were identified by searching the resulting peak lists against the Uniprot database (Homo sapiens), with exclusion of common contaminants. Because glucose is not used for amino acid production and glycosylated peptides were not measured, in vivo 2H labeling should not have affected protein quantifications. All results have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository (data set identifier PXD001674; DOI: 10.6019/PXD001674).34
Hierarchical clustering
Hierarchical clustering was employed to quantify the dissimilarities between the neutrophil subsets and the individual samples. Proteins detected or reliably quantified in only 1 of 3 individuals in any of the subsets were excluded from analysis. Remaining missing values were mean imputed. Using the pvclust plugin (version 1.3-0) for R (version 3.1.1), a Euclidean dissimilarity matrix was calculated from protein abundances and clustered hierarchically using the averages method. Significance of the resulting clusters was tested with multiscale bootstrapping (10 000 repeats) and reported as approximate unbiased P values ± 95% CIs of the P values.35
Clustering of protein expression data
Sums of protein quantifications from 3 individual samples were standardized to an average expression of 0, with a standard deviation of 1. Proteins with missing values for ≥2 individuals in any of the 3 subsets were excluded from analysis. Standardized expression profiles were subjected to Fuzzy C-means clustering using R with the Mfuzz plugin (version 2.3.1).36 In addition to assigning each protein to a cluster, fuzzy clustering indicates how well each protein fits in each cluster. By choosing a minimal required membership value, outliers are excluded from analysis, making fuzzy clustering preferable over hard clustering methods such as the k-means method.
The required number of clusters was determined by calculating the average centroid distance for clustering runs with 2 up to 40 clusters.37 After 7 clusters, addition of extra clusters hardly improved the clustering results. Thus, the number of clusters was set to 7. To prevent clustering of random data while keeping the false-negative rate as low as possible, the optimal value for fuzzifier parameter m was calculated as described previously,37 resulting in a value of 3.69.
GO enrichment analysis
Proteins with cluster membership values of at least 0.33 were analyzed by gene ontology (GO) enrichment analysis. Gene names of identified proteins were obtained from the Uniprot database. Subsequently, GO terms of biological processes, molecular functions, and cellular components were obtained for each protein and analyzed using the GO enrichment analysis and visualization tool (GOrilla; accessed 20 January 2015).38,39 Enrichment was determined for proteins in each cluster compared with the background set of all identified proteins using an exact mHG P value computation, corrected for multiplicity using the Benjamini and Hochberg40 false-discovery rate correction.38 Because this resulted in a list of up to 120 significantly enriched GO terms in a cluster, results were summarized by removing highly similar GO terms with REViGO and visualized in R using the ggplot2 plugin (version 0.9.3.1).41
Statistics on clinical data
Clinical data were tested for significant deviation from t = 0 using 1-way repeated-measures analysis of variance, with P values corrected for multiplicity using Dunnett’s correction.
Results
Kinetics of clinical and inflammatory parameters after endotoxin challenge
As shown previously,42 infusion of LPS (2 ng/kg) in healthy volunteers induced clinical symptoms reminiscent of systemic inflammatory response syndrome. Disease scores were increased 1.5 hours after LPS injection, followed by an increase in heart rate and body temperature at 2 to 5.5 hours post-LPS (supplemental Figure 3). The mean arterial pressure decreased from 2 hours after LPS injection until the end of the experiment.
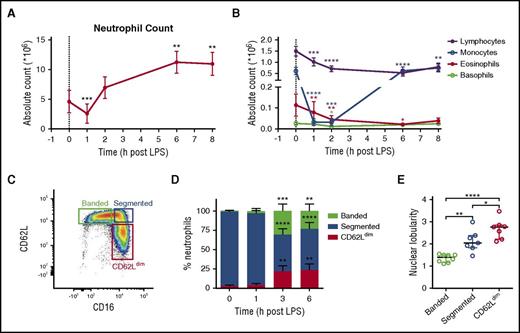
Blood counts of all leukocytes were decreased 1 hour after LPS injection (Figure 1A-B). Circulating monocyte numbers had normalized after 6 hours, whereas lymphocyte, eosinophil, and basophil numbers remained low until 8 hours after LPS injection. After an initial decrease in neutrophil numbers at 1 hour after LPS, numbers increased to levels 2.5-fold higher than the pre-LPS count. FACS analysis revealed 3 neutrophil subsets based on CD16/CD62L expression (Figure 1C), which were present at 3 and 6 hours post-LPS challenge (Figure 1D). Microscopic examination of sorted subsets showed an increased average number of nuclear lobes in the CD16bright/CD62Ldim population and a banded nuclear phenotype for the CD16dim/CD62Lbright population (Figure 1E).11
Kinetics of white blood cells and neutrophil subsets after LPS injection. (A) Neutrophil counts were decreased 1 hour after LPS injection (t = 0), followed by increased counts. (B) Counts of other leukocytes were also decreased at t = 1 hour, but most had recovered by t = 6 or 8 hours. (C) Different neutrophil subsets were identified (t = 3 hours) based on CD16 and CD62L expression. (D) The appearance of the banded and CD62Ldim subsets was followed over time. (E) After sorting, microscopic analysis of the different subsets isolated at t = 3 hours post–LPS challenge revealed an increasing nuclear lobularity from banded to segmented to CD62Ldim neutrophils. Symbols and bars represent mean ± 95% CI (n = 20) (A,B,D) or medians ± interquartile range (n = 7-8) (E). *P < .05, **P < .01, ***P < .001, and ****P < .0001 compared with t = 0 hours, as determined by repeated-measures 1-way analysis of variance with Dunnett’s or Holm-Sidak correction for multiplicity where applicable.
Kinetics of white blood cells and neutrophil subsets after LPS injection. (A) Neutrophil counts were decreased 1 hour after LPS injection (t = 0), followed by increased counts. (B) Counts of other leukocytes were also decreased at t = 1 hour, but most had recovered by t = 6 or 8 hours. (C) Different neutrophil subsets were identified (t = 3 hours) based on CD16 and CD62L expression. (D) The appearance of the banded and CD62Ldim subsets was followed over time. (E) After sorting, microscopic analysis of the different subsets isolated at t = 3 hours post–LPS challenge revealed an increasing nuclear lobularity from banded to segmented to CD62Ldim neutrophils. Symbols and bars represent mean ± 95% CI (n = 20) (A,B,D) or medians ± interquartile range (n = 7-8) (E). *P < .05, **P < .01, ***P < .001, and ****P < .0001 compared with t = 0 hours, as determined by repeated-measures 1-way analysis of variance with Dunnett’s or Holm-Sidak correction for multiplicity where applicable.
In vivo 2H labeling
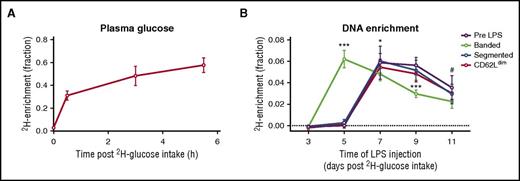
Because neutrophil progenitors do not proliferate after the myelocyte stage, in vivo 6,6-2H2-glucose labeling can be used to determine the time required for the different subsets to mature and be released into the bloodstream. In contrast to deuterium labels such as 2H2O, 6,6-2H2-glucose has a rapid turnover in the circulation (Figure 2A) and is rapidly metabolized. This allows the application of a short labeling pulse followed by a chase experiment. To correct for differences in label uptake between volunteers, the availability of label was determined in the plasma of each volunteer (Figure 2A). Corrected DNA 2H kinetcs under homeostatic conditions were determined on neutrophils isolated before injection of LPS. In these cells, the first 2H was detected in peripheral blood neutrophil DNA at day 7 after intake of label (Figure 2B). This is in line with the notion that conventional neutrophils mature approximately 6 days in the bone marrow after the last division of their progenitors (myelocytes).29 Segmented neutrophils isolated at 3 hours post-LPS injection showed 2H-enrichment curves similar to neutrophils isolated under homeostatic conditions, suggesting that the maturation kinetics of segmented neutrophils were not affected by LPS. CD62Ldim neutrophils also showed similar 2H-enrichment kinetics to homeostatic neutrophils, with the highest enrichment at 7 days after intake of label. In contrast, the banded neutrophil subset showed a different DNA 2H enrichment, with highest enrichment measured as early as 5 days postlabel intake, suggesting that maturation into a banded cell takes approximately 2 days less than maturation into the 2 other phenotypes. No significant differences in labeling kinetics were observed between placebo- and C1 inhibitor–treated groups (supplemental Figure 4). Thus, labeling could be combined from both treatment groups, providing additional statistical power.
Label kinetics in plasma and in DNA of neutrophil subsets. Healthy volunteers were labeled with 6,62H2-glucose over the course of 6 hours. (A) During this period, the availability of label was monitored by measuring the fraction of 6,6-2H2-glucose in the total blood glucose pool in blood samples obtained by skin pricks. (B) DNA 2H enrichment of the different inflammatory neutrophil subsets 3 hours after LPS injection or total neutrophils isolated 5 minutes pre-LPS injection at 3 to 11 days after 6,6-2H2-glucose. Results suggest that banded neutrophils are less mature than the 2 other subsets. (A) Fraction enrichment was corrected for availability of label for each individual. Symbols indicate mean ± standard deviation, with (A) n = 20 and (B) n = 4 volunteers at each time point, with a total of 20 volunteers. *P < .05 for banded compared with segmented neutrophils, #P < .05 for banded neutrophils compared with neutrophils isolated before LPS injection, and ***P < .001 for banded neutrophils compared with all 3 other subsets as determined by a repeated-measures 2-way analysis of variance with Tukey’s correction for multiplicity.
Label kinetics in plasma and in DNA of neutrophil subsets. Healthy volunteers were labeled with 6,62H2-glucose over the course of 6 hours. (A) During this period, the availability of label was monitored by measuring the fraction of 6,6-2H2-glucose in the total blood glucose pool in blood samples obtained by skin pricks. (B) DNA 2H enrichment of the different inflammatory neutrophil subsets 3 hours after LPS injection or total neutrophils isolated 5 minutes pre-LPS injection at 3 to 11 days after 6,6-2H2-glucose. Results suggest that banded neutrophils are less mature than the 2 other subsets. (A) Fraction enrichment was corrected for availability of label for each individual. Symbols indicate mean ± standard deviation, with (A) n = 20 and (B) n = 4 volunteers at each time point, with a total of 20 volunteers. *P < .05 for banded compared with segmented neutrophils, #P < .05 for banded neutrophils compared with neutrophils isolated before LPS injection, and ***P < .001 for banded neutrophils compared with all 3 other subsets as determined by a repeated-measures 2-way analysis of variance with Tukey’s correction for multiplicity.
Hierarchical clustering of neutrophil subsets based on protein expression data
Proteome analysis by LC-MS/MS identified and quantified 2161 unique proteins in the 3 neutrophil subsets obtained from 3 individuals 3 hours after LPS injection (supplemental Table 2). LC-MS/MS protein quantification was validated for several proteins with different expression patterns across the 3 subsets (supplemental Figure 2) by western blotting or by comparison with previously published FACS data.11 All tested proteins showed highly similar expression patterns with the different techniques, even though results obtained by western blotting showed a larger variability than those obtained by LC-MS/MS.
The relations between the 3 neutrophil subsets and each replicate were assessed using hierarchical clustering on proteins with no more than 1 missing value in each of the subsets (Figure 3; 1755 of 2161 proteins). For hierarchical clustering, a Euclidean dissimilarity matrix was created to determine the correlation between all samples. Subsequently, a dendrogram was created to visualize the relations between samples (Figure 3B). Replicates of each subset clustered tightly together in this dendrogram. Using multiscale bootstrapping, we determined that the existence of these clusters was supported by our data with a probability of >99.9%.35 At the proteome level, banded and segmented neutrophils correlated better and were distinctly separated from the CD62Ldim neutrophils. Use of normalized protein expressions or different distance matrix calculations did not result in formation of different clusters (data not shown).
Hierarchical clustering of protein expression data. (A) Heatmap of normalized protein expression profiles. (B) Hierarchical clustering of non-normalized protein expression using averages clustering on a Euclidean distance matrix. Clusters were tested for statistical significance using multiscale bootstrapping, which revealed 4 statistically significant clusters (indicated by red boxes): 1 for each neutrophil subset and 1 for the banded and segmented neutrophils together. P values are approximately unbiased (AU; ie, an AU P value of .95 indicates that the existence of 2 clusters is supported by the data with 95% confidence). Numbers between brackets indicate the standard error of the AU P value estimation.
Hierarchical clustering of protein expression data. (A) Heatmap of normalized protein expression profiles. (B) Hierarchical clustering of non-normalized protein expression using averages clustering on a Euclidean distance matrix. Clusters were tested for statistical significance using multiscale bootstrapping, which revealed 4 statistically significant clusters (indicated by red boxes): 1 for each neutrophil subset and 1 for the banded and segmented neutrophils together. P values are approximately unbiased (AU; ie, an AU P value of .95 indicates that the existence of 2 clusters is supported by the data with 95% confidence). Numbers between brackets indicate the standard error of the AU P value estimation.
Characterization of neutrophil subsets by proteome analysis
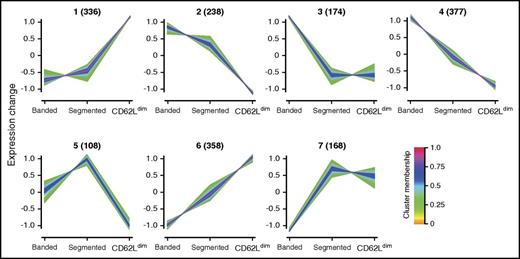
To further examine the relation between the 3 neutrophil subsets, protein expression profiles were examined using functional enrichment analysis. To this end, proteins were first clustered based on their relative expression between the 3 subsets. Before clustering, the optimal number of protein clusters was determined to be 7, because addition of extra clusters did not lead to better results. The resulting protein clusters were clearly different in their relative expression between the 3 neutrophil subsets (Figure 4). A total of 618 proteins had an expression pattern that differed too much from the 7 clusters to be assigned to any of the clusters and were treated as a separate cluster.
Clustering of protein expression profiles. Clustering analysis divides normalized protein expression patterns into 7 distinct clusters. Three clusters have high expression in 1 subset compared with the others (clusters 1, 3, and 5 for CD62Ldim, banded, and segmented neutrophils, respectively), 2 clusters have decreased expression in 1 subset (clusters 2 and 7 for CD62Ldim and banded neutrophils, respectively), and 2 clusters have gradually increasing (cluster 6) or decreasing (cluster 4) protein expression levels from banded to segmented and CD62Ldim. Each line represents the expression pattern of 1 protein. Line colors indicate the cluster membership value (ie, the higher the membership value, the more similar to the cluster average). A minimum cluster membership value of 0.33 was used for further analysis. Numbers between brackets indicate the number of proteins in each cluster with cluster membership values above 0.33.
Clustering of protein expression profiles. Clustering analysis divides normalized protein expression patterns into 7 distinct clusters. Three clusters have high expression in 1 subset compared with the others (clusters 1, 3, and 5 for CD62Ldim, banded, and segmented neutrophils, respectively), 2 clusters have decreased expression in 1 subset (clusters 2 and 7 for CD62Ldim and banded neutrophils, respectively), and 2 clusters have gradually increasing (cluster 6) or decreasing (cluster 4) protein expression levels from banded to segmented and CD62Ldim. Each line represents the expression pattern of 1 protein. Line colors indicate the cluster membership value (ie, the higher the membership value, the more similar to the cluster average). A minimum cluster membership value of 0.33 was used for further analysis. Numbers between brackets indicate the number of proteins in each cluster with cluster membership values above 0.33.
Subsequently, GO enrichment analysis was performed on the 7 clusters of proteins and the cluster of nonclassified proteins. GO terms describe biological processes, molecular functions, or cellular components and are annotated to genes/proteins based on evidence from literature. GO enrichment analysis revealed significant GO-term enrichments in clusters 1, 2, 4, and 6. (Table 1 lists the 10 most significantly enriched GO terms in each cluster, and supplemental Table 3 provides the complete list of enrichments).
Top 10 most significantly enriched GO terms per cluster
| Cluster . | FDR-corrected P . | Enrichment . | Type . |
|---|---|---|---|
| Cluster 1: CD62Ldim ↑ | |||
| Extracellular space | 4.98E-09 | 2.43 | Component |
| Extracellular region | 1.98E-08 | 2.69 | Component |
| Plasma membrane | 5.60E-06 | 1.58 | Component |
| Response to organic substance | 1.08E-05 | 1.86 | Function |
| Defense response | 1.40E-05 | 1.86 | Function |
| Response to external stimulus | 1.75E-05 | 2.07 | Function |
| Response to oxygen-containing compound | 2.22E-05 | 2.04 | Function |
| Response to external biotic stimulus | 2.41E-05 | 2.25 | Function |
| Response to chemical | 2.52E-05 | 1.69 | Function |
| Response to biotic stimulus | 3.41E-05 | 2.22 | Function |
| Cluster 2: CD62Ldim↓ | |||
| Ribonucleoprotein complex | 3.72E-07 | 2.25 | Component |
| Nucleic acid binding | 1.01E-05 | 1.61 | Function |
| RNA binding | 2.02E-05 | 1.66 | Function |
| RNA metabolic process | 4.30E-05 | 1.82 | Process |
| Poly(A) RNA binding | 9.61E-05 | 1.7 | Function |
| Nucleic acid metabolic process | 3.81E-04 | 1.67 | Process |
| Structural constituent of ribosome | 9.23E-04 | 2.99 | Function |
| Ribosomal subunit | 1.55E-03 | 2.91 | Component |
| mRNA metabolic process | 1.82E-03 | 2.04 | Process |
| Translational elongation | 2.04E-03 | 2.89 | Process |
| Cluster 4: band > segmented > CD62Ldim | |||
| Ribonucleoprotein complex | 1.07E-15 | 2.34 | Component |
| Ribosomal subunit | 2.57E-13 | 3.55 | Component |
| RNA metabolic process | 4.25E-13 | 1.89 | Process |
| Cotranslational protein targeting to membrane | 4.97E-13 | 3.32 | Process |
| SRP-dependent cotranslational protein targeting to membrane | 5.26E-13 | 3.36 | Process |
| Protein targeting to ER | 7.39E-13 | 3.28 | Process |
| Nucleic acid metabolic process | 7.50E-13 | 1.78 | Process |
| Translational initiation | 9.36E-13 | 3.03 | Process |
| Protein localization to ER | 1.36E-12 | 3.2 | Process |
| Establishment of protein localization to ER | 1.54E-12 | 3.2 | Process |
| Cluster 6: CD62Ldim> segmented > band | |||
| Extracellular vesicular exosome | 2.32E-04 | 1.26 | Component |
| Extracellular vesicle | 2.90E-04 | 1.26 | Component |
| Extracellular membrane-bounded organelle | 3.86E-04 | 1.26 | Component |
| Vesicle | 5.18E-04 | 1.23 | Component |
| Extracellular region part | 5.57E-04 | 1.24 | Component |
| Extracellular organelle | 5.80E-04 | 1.26 | Component |
| Membrane-bounded vesicle | 8.88E-04 | 1.25 | Component |
| Actin binding | 7.31E-03 | 2.18 | Function |
| Negative regulation of Wnt signaling pathway | 2.00E-02 | 2.45 | Process |
| Nicotinamide nucleotide metabolic process | 2.02E-02 | 3.72 | Process |
| Cluster . | FDR-corrected P . | Enrichment . | Type . |
|---|---|---|---|
| Cluster 1: CD62Ldim ↑ | |||
| Extracellular space | 4.98E-09 | 2.43 | Component |
| Extracellular region | 1.98E-08 | 2.69 | Component |
| Plasma membrane | 5.60E-06 | 1.58 | Component |
| Response to organic substance | 1.08E-05 | 1.86 | Function |
| Defense response | 1.40E-05 | 1.86 | Function |
| Response to external stimulus | 1.75E-05 | 2.07 | Function |
| Response to oxygen-containing compound | 2.22E-05 | 2.04 | Function |
| Response to external biotic stimulus | 2.41E-05 | 2.25 | Function |
| Response to chemical | 2.52E-05 | 1.69 | Function |
| Response to biotic stimulus | 3.41E-05 | 2.22 | Function |
| Cluster 2: CD62Ldim↓ | |||
| Ribonucleoprotein complex | 3.72E-07 | 2.25 | Component |
| Nucleic acid binding | 1.01E-05 | 1.61 | Function |
| RNA binding | 2.02E-05 | 1.66 | Function |
| RNA metabolic process | 4.30E-05 | 1.82 | Process |
| Poly(A) RNA binding | 9.61E-05 | 1.7 | Function |
| Nucleic acid metabolic process | 3.81E-04 | 1.67 | Process |
| Structural constituent of ribosome | 9.23E-04 | 2.99 | Function |
| Ribosomal subunit | 1.55E-03 | 2.91 | Component |
| mRNA metabolic process | 1.82E-03 | 2.04 | Process |
| Translational elongation | 2.04E-03 | 2.89 | Process |
| Cluster 4: band > segmented > CD62Ldim | |||
| Ribonucleoprotein complex | 1.07E-15 | 2.34 | Component |
| Ribosomal subunit | 2.57E-13 | 3.55 | Component |
| RNA metabolic process | 4.25E-13 | 1.89 | Process |
| Cotranslational protein targeting to membrane | 4.97E-13 | 3.32 | Process |
| SRP-dependent cotranslational protein targeting to membrane | 5.26E-13 | 3.36 | Process |
| Protein targeting to ER | 7.39E-13 | 3.28 | Process |
| Nucleic acid metabolic process | 7.50E-13 | 1.78 | Process |
| Translational initiation | 9.36E-13 | 3.03 | Process |
| Protein localization to ER | 1.36E-12 | 3.2 | Process |
| Establishment of protein localization to ER | 1.54E-12 | 3.2 | Process |
| Cluster 6: CD62Ldim> segmented > band | |||
| Extracellular vesicular exosome | 2.32E-04 | 1.26 | Component |
| Extracellular vesicle | 2.90E-04 | 1.26 | Component |
| Extracellular membrane-bounded organelle | 3.86E-04 | 1.26 | Component |
| Vesicle | 5.18E-04 | 1.23 | Component |
| Extracellular region part | 5.57E-04 | 1.24 | Component |
| Extracellular organelle | 5.80E-04 | 1.26 | Component |
| Membrane-bounded vesicle | 8.88E-04 | 1.25 | Component |
| Actin binding | 7.31E-03 | 2.18 | Function |
| Negative regulation of Wnt signaling pathway | 2.00E-02 | 2.45 | Process |
| Nicotinamide nucleotide metabolic process | 2.02E-02 | 3.72 | Process |
ER, endoplasmic reticulum; FDR, false-discovery rate; SRP, signal recognition particle.
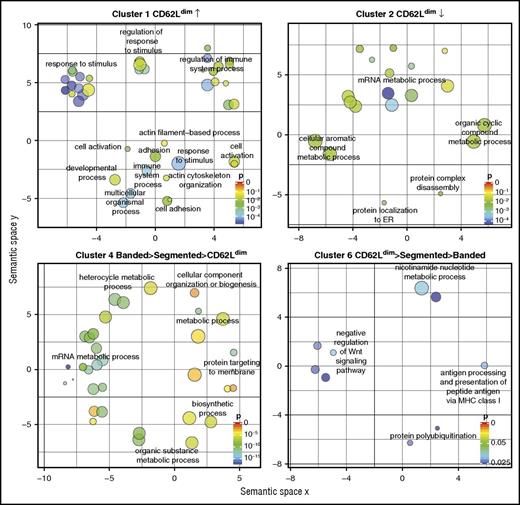
Using ReViGO, highly similar GO terms were removed from clusters with significant enrichments, and remaining GO terms for biological processes were clustered based on semantic similarity (Figure 5). Thereby, enrichments of protein functions could clearly be recognized. Cluster 1, which consisted of 336 proteins upregulated in CD62Ldim neutrophils, contained genes involved in adhesion and activation, response to stimuli, regulation of these responses, and regulation of the immune system in general. Cluster 2, consisting of 238 proteins with a lower expression in CD62Ldim neutrophils, showed enrichment of proteins involved in protein shuttling, ribosomal proteins, and proteins involved in RNA metabolism, splicing, and translation. Similarly, cluster 4, which contained 337 proteins gradually reduced from banded via segmented to CD62Ldim neutrophils, consisted mostly of proteins involved in mRNA processing and biosynthesis. Lastly, cluster 6 contained 358 proteins with a lower expression in banded neutrophils, compared with segmented and CD62Ldim neutrophils. This cluster showed enrichment in proteins involved in nucleotide metabolism, major histocompatibility complex class I antigen presentation, protein degradation, and suppression of the Wnt signaling pathway. Taken together, these data implicate differential functionalities for the different neutrophil subsets.
GO enrichment analysis for protein involvement in cellular processes. The 7 clusters from Figure 4 were subjected to GO enrichment analysis. After correction for multiple testing, 4 clusters revealed significant GO enrichments, which are plotted based on semantic similarities (ie, similar terms grouped together). Colors indicate false-discovery rate P values for each term, and the size indicates how general a term is, with a smaller size for a more specific term. Highly similar terms were removed for clarity but are provided in supplementary Table 3. ER, endoplasmic reticulum; MHC, major histocompatibility complex.
GO enrichment analysis for protein involvement in cellular processes. The 7 clusters from Figure 4 were subjected to GO enrichment analysis. After correction for multiple testing, 4 clusters revealed significant GO enrichments, which are plotted based on semantic similarities (ie, similar terms grouped together). Colors indicate false-discovery rate P values for each term, and the size indicates how general a term is, with a smaller size for a more specific term. Highly similar terms were removed for clarity but are provided in supplementary Table 3. ER, endoplasmic reticulum; MHC, major histocompatibility complex.
Discussion
Our data challenge the hypothesis that the different neutrophil subsets (banded, segmented, and CD62Ldim cells) follow a linear differentiation from banded (immature) to segmented (mature) to CD62Ldim (aged), as is the consensus in the field.4,11,12 After pulse-chase labeling with 6,6-2H2-glucose, labeled banded neutrophils were observed several days earlier in the blood than labeled segmented and CD62Ldim neutrophils. Because 2H labeling of DNA only takes place during the S phase of the cell cycle, these data demonstrate that banded neutrophils observed in acute inflammation take less time to mature from myelocytes than the other 2 subsets and are therefore likely to be cells prematurely mobilized from the bone marrow.43,44 These data are in agreement with those of previous studies in patients with cancer.15,16 This mobilization is generally thought to be a compensation of the bone marrow to release as many neutrophils as possible, even if some of those are not yet fully mature and might have a low capacity for clearing pathogens.45 Functional enrichment analysis supported the hypothesis that banded neutrophils are immature cells still in the process of producing large amounts of proteins, because these cells showed the highest expression of proteins involved in de novo protein synthesis (Figure 5; supplemental Figure 5). CD62Ldim neutrophils, conversely, showed enrichment of proteins involved in immune regulation, which is in line with their previously published capacity for suppressing T-cell–suppressing proliferation.11 It is unknown, however, whether the capacity of CD62Ldim neutrophils for suppressing T-cell proliferation is in any way related to their low capacity for protein production.
In contrast to the banded neutrophils, CD62Ldim neutrophils had kinetics similar to those of segmented neutrophils, with respect to maturation time and disappearance rates from peripheral blood. This suggests that CD62Ldim neutrophils need the same time to mature as segmented neutrophils and are thus not simply aged segmented neutrophils. These results are in agreement with a previous study describing the maturation time of CD62Ldim neutrophils in a patient with pernicious anemia.16 It should be noted, however, that the hypersegmentation observed in pernicious anemia is caused by defects in the DNA replication machinery,46 which causes banded neutrophils to immediately mature into hypersegmented neutrophils.18 Because the maturation time of CD62Ldim neutrophils after the last DNA synthesis was at least 5 days, it is unlikely that the DNA replication defects causing hypersegmentation in pernicious anemia caused the hypersegmentation in the cells observed 3 hours after acute inflammation. The highly similar kinetics of segmented and CD62Ldim neutrophils were not reflected by comparable protein expression profiles. Hierarchical clustering of the proteomes of the different subsets revealed the segmented subset to be more similar to the less mature banded neutrophils than to the CD62Ldim neutrophils. Thus, the difference between segmented and CD62Ldim neutrophils is larger than the difference caused by 2 days of maturation from banded to segmented neutrophils.
The large difference in the proteomes of CD62Ldim neutrophils compared with the other 2 subsets raises the issue of the putative origin of these cells. It is less likely that the CD62Ldim neutrophil subset originates from mature neutrophils in response to LPS. Although it has been shown that neutrophils isolated from healthy individuals can produce novel proteins after incubation with high doses of LPS in vitro (100 ng/mL),27 this did not seem to occur in our study for various reasons. In vitro experiments showing LPS-induced neutrophil activation use doses in the upper nanomolar range, whereas the dose used in our study was 2 ng/kg body weight (∼50 pg/mL plasma).47 To our knowledge, no studies have shown neutrophils to be directly responsive in the lower picomolar range. This consideration is in line with the results coming from the comparison between our data with the proteomes published by Fessler et al,47 who studied LPS-induced changes in neutrophils activated in vitro. Only 3 of 18 differentially expressed proteins found in neutrophils activated by LPS in vitro showed behavior similar to that seen in our proteome profiling, whereas an additional 4 proteins showed the opposite behavior (supplemental Table 1). This makes it unlikely that CD62Ldim neutrophils are generated in direct response to LPS stimulation. In addition, all proteins involved in transcription, translation, and mRNA/protein transport that we detected in our proteome analysis were least abundant in CD62Ldim neutrophils (supplemental Figure 5). This indicates that CD62Ldim neutrophils have an even lower capacity for protein production than segmented neutrophils, while having a higher expression of 336 proteins compared with segmented and banded cells (Figure 4, cluster 1). Therefore, we propose that the CD62Ldim neutrophil phenotype is not rapidly induced from normal neutrophils in response to LPS but instead is a neutrophil subset that enters the bloodstream in response to inflammation.
The existence of neutrophil subsets residing outside of the circulation is supported by the fact that under homeostasis, large numbers of neutrophils do not reside in the peripheral blood. Neutrophils have been found in the lungs, spleen, liver, and bone marrow or can be recruited from the marginated pool.6,48,49 In addition, reverse transmigrated neutrophils have been described that return from the tissue back into the bloodstream.50 It is yet to be determined which tissue sites are the source of the neutrophil subsets released during acute inflammation. Reverse transmigrated neutrophils are not a likely source of CD62Ldim neutrophils, because CD62Ldim neutrophils do not exhibit the profile of reverse migrated cells: no increased expression of ICAM-1 (CD54) and no decreased CXCR1 or CXCR2 expression as determined by flow cytometric analysis11 as well as by proteomics (supplemental Table 2). Also, the high expression of CD11c on CD62Ldim neutrophils is not found on reverse migrated neutrophils.50 The marginated pool does not seem a likely source of CD62Ldim neutrophils either, because it consists of neutrophils with a similar phenotype as those in the circulation.5,49,51 In contrast, the increase in neutrophil numbers after LPS results from the entrance of cells into the bloodstream with different characteristics than those present in homeostasis.5
CD62Ldim neutrophils have been described in the bone marrow, albeit at lower numbers than banded and segmented neutrophils.20 Therefore, the bone marrow is also a potential source of CD62Ldim neutrophils. Alternatively, neutrophil reserves have been described in the liver and spleen, and these are therefore also potential organs from which CD62Ldim neutrophils are recruited after LPS injection.6,48,49 The kinetics of 2H-glucose labeling depend on both blood and bone marrow kinetics.52,53 The fact that mature and CD62Ldim neutrophils show similar label kinetics suggests a shared progenitor and similar kinetics in bone marrow and blood for the 2 cell types, but the exact developmental relation between the subsets remains to be elucidated.
Taken together, our study shows that CD62Ldim neutrophils detected in the bloodstream after LPS challenge are of similar age as segmented neutrophils but have a proteome profile that places them apart from both segmented and the 2-day-younger banded neutrophils. Therefore, we propose CD62Ldim is a subset of neutrophils that are only recruited to the bloodstream during acute inflammatory conditions, where they can engage in immune regulation to fine tune acute immune responses.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Sigrid Otto for her help with the gas chromatography mass spectrometry, the fluorescence-activated cell sorter operators of the Laboratory for Translational Immunology flow core, and the research nurses of the Radboud Medical Centre for taking care of our volunteers.
This work was supported by the Dutch Lung Foundation (grant 3.2.10.052) and Sanquin Blood Supply. The proteome analysis was supported by the project Proteins At Work (project 184.032.201), a program of the Netherlands Proteomics Centre financed by the Netherlands Organization for Scientific Research as part of the National Roadmap Large-Scale Research Facilities of the Netherlands.
The funders had no role in study design, data collection or analysis, decision to publish, or preparation of the manuscript.
Authorship
Contribution: T.T. isolated cells, analyzed samples for 2H enrichment, and performed all statistical analyses; P.W. performed protein identification and quantifications, supervised by A.S. and A.J.R.H. T.T. performed analysis of the proteomics data, supervised by A.S. and A.J.R.H.; M.H. was responsible for volunteer screenings and lipopolysaccharide injections and collected clinical data; M.H. and T.T. performed 6,6-2H2 labeling; A.J.R.H., A.S., J.A.M.B., K.T., L.P.L., P.P., N.V., and L.K. participated in the design, interpretation, and coordination of the study; T.T. wrote the first draft of the manuscript, which was revised with the help of all other authors; and all authors read and approved the final manuscript.
Conflict-of-interest disclosure: A.S. is currently employed by Janssen, Pharmaceutical Companies of Johnson & Johnson. The work described in this manuscript was entirely executed before and without affiliation with the work of A.S. at Janssen. As such there is no relationship at all between this commercial company and the currently presented study. The remaining authors declare no competing financial interests.
Correspondence: Leo Koenderman, HP. E 03.511, Heidelberglaan 100, 3584CX Utrecht, The Netherlands; e-mail: l.koenderman@umcutrecht.nl.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal