Key Points
iNKT cells control CLL progression in both mice and patients and this inversely correlates with CD1d expression by leukemia cells.
Human iNKT cells indirectly hinder CLL survival by restraining proleukemia monocyte-derived nurse-like cells.
Abstract
Chronic lymphocytic leukemia (CLL) is characterized by the expansion of malignant CD5+ B lymphocytes in blood, bone marrow, and lymphoid organs. CD1d-restricted invariant natural killer T (iNKT) cells are innate-like T lymphocytes strongly implicated in tumor surveillance. We investigated the impact of iNKT cells in the natural history of the disease in the Eμ-Tcl1 (Tcl1) CLL mouse model and 68 CLL patients. We found that Tcl1-CLL cells express CD1d and that iNKT cells critically delay disease onset but become functionally impaired upon disease progression. In patients, disease progression correlates with high CD1d expression on CLL cells and impaired iNKT cells. Conversely, disease stability correlates with negative or low CD1d expression on CLL cells and normal iNKT cells, suggesting indirect leukemia control. iNKT cells indeed hinder CLL survival in vitro by restraining CD1d-expressing nurse-like cells, a relevant proleukemia macrophage population. Multivariable analysis identified iNKT cell frequency as an independent predictor of disease progression. Together, these results support the contribution of iNKT cells to CLL immune surveillance and highlight iNKT cell frequency as a prognostic marker for disease progression.
Introduction
Chronic lymphocytic leukemia (CLL) is the most common adult leukemia,1,2 characterized by the expansion of mature malignant CD5+ B cells.3 CLL is clinically heterogeneous, and patients experience either stable or progressive disease with accumulation of malignant B cells in lymph nodes, spleen, bone marrow, and peripheral blood.4 Nurse-like cells (NLCs) are CLL-specific tumor-associated macrophages (TAMs), which can differentiate in vitro from monocytes cultured with CLL cells.5 They sustain malignant B-cell survival6,7 and are a major cellular component of the leukemia microenvironment in lymphoid tissues8,9 of patients with progressive but not stable disease, suggesting that these cells have an impact on the natural history of CLL.10,11
Type I invariant natural killer T (iNKT) cells are a conserved T-cell subset expressing a semi-invariant T-cell receptor (TCR) containing the Vα14-Jα18 and Vα24-Jα18 chains in mice and humans, respectively. iNKT cells react with self- or microbial-derived lipid antigens, such as α-galactosyl ceramide (α-GalCer), presented by the HLA class I–related molecule CD1d. iNKT cells exhibit an innate-like effector phenotype and swiftly respond to stimulation by upregulating costimulatory molecules and cytokines, conferring on them a central role in tumor immune surveillance.12 iNKT cells can control tumor growth either by direct recognition of CD1d-expressing tumor cells, or indirectly, by activation of antitumor CD8+ T and NK cells or by modulation of protumor myelomonocytic cells.13-18 In cancer patients, decreased iNKT cell numbers and interferon-γ (IFN-γ) production correlate with adverse prognosis.19-21
In CLL patients, it was reported that CD1d expression by malignant cells correlates with adverse prognosis, whereas iNKT cells can recognize leukemia cells in vitro when preloaded with synthetic α-GalCer.22-24 However, the role of iNKT cells in the natural history of CLL is essentially unknown.
Our study addresses this issue in Tcl1 mice, a widely accepted mouse model of CLL,25 and in a cohort of 68 CLL patients. In mice, we found that iNKT cells controlled leukemia onset but became functionally impaired upon disease progression. iNKT cells were consistently deeply impaired in patients with disease progression, and this somewhat unexpectedly correlates with high CD1d expression on autologous CLL cells. By contrast, in patients with stable disease, CLL cells display no or low CD1d expression, whereas iNKT cells retain intact functions and can control leukemia cell survival in vitro by restraining protumor functions of NLCs. iNKT cell frequency has emerged as a new valuable prognostic factor for progressive leukemia.
Methods
Mice
C57BL/6N wild-type (wt) (Charles River Laboratories, Calco, Italy), Eμ-TCL1,25 Jα18−/−,26 and CD1d−/− mice27 (all on the C57BL/6N background) were treated in accordance with the Institutional Animal Care and Use Committee of San Raffaele Scientific Institute, Milan, Italy. Eμ-Tcl1 mice were crossed with Jα18−/− or CD1d−/− mice to obtain Tcl1-Jα18−/− or Tcl1-CD1d−/− mice.
Patients
CLL diagnosis, prognostic factors, and disease progression (Table 1) were documented according to the International Workshop on CLL 2008 criteria (IWCLL 2008).28 All patients were either untreated or off therapy for at least 6 months (range, 6 to 12 months) before the beginning of the study. Patients with progressive disease who were receiving antileukemia treatment were excluded from the study. Progressive disease was defined per IWCLL 2008 criteria and was based on the presence of at least 1 of the following clinical parameters: anemia and/or thrombocytopenia as a result of bone marrow infiltration by CLL, bulky symptomatic or progressive lymph nodes, massive symptomatic or progressive splenomegaly, uncontrolled autoimmune anemia and/or thrombocytopenia, lymphocyte doubling time <6 months, and/or B symptoms (drenching night sweats, fever, unexplained weight loss). Peripheral blood from CLL patients and age-matched healthy donors and lymph node samples from CLL patients were obtained after patients provided written informed consent in accordance with the Declaration of Helsinki.
Patient characteristics
| Characteristic . | No. of patients/total patients . | % . |
|---|---|---|
| Patient parameters | ||
| Total no. of patients | 68 | |
| Median age at diagnosis (range), y | 67 (41-89) | |
| Male sex | 39 | |
| Clinical parameters | ||
| Rai stage at diagnosis | ||
| 0 | 42/60 | 70 |
| 1-4 | 18/60 | 30 |
| IGHV mutational status | ||
| Mutated | 37/58 | 63.8 |
| Unmutated | 21/58 | 36.2 |
| CD38 expression, % | ||
| <30 | 53/65 | 81.5 |
| >30 | 12/65 | 18.5 |
| ZAP-70 expression, % | ||
| <20 | 34/59 | 57.6 |
| >20 | 25/59 | 42.4 |
| Chromosomal aberrations | ||
| Trisomy 12 | 7/63 | 11.1 |
| FISH del11q | 10/63 | 15.9 |
| FISH del17p | 11/63 | 17.4 |
| Characteristic . | No. of patients/total patients . | % . |
|---|---|---|
| Patient parameters | ||
| Total no. of patients | 68 | |
| Median age at diagnosis (range), y | 67 (41-89) | |
| Male sex | 39 | |
| Clinical parameters | ||
| Rai stage at diagnosis | ||
| 0 | 42/60 | 70 |
| 1-4 | 18/60 | 30 |
| IGHV mutational status | ||
| Mutated | 37/58 | 63.8 |
| Unmutated | 21/58 | 36.2 |
| CD38 expression, % | ||
| <30 | 53/65 | 81.5 |
| >30 | 12/65 | 18.5 |
| ZAP-70 expression, % | ||
| <20 | 34/59 | 57.6 |
| >20 | 25/59 | 42.4 |
| Chromosomal aberrations | ||
| Trisomy 12 | 7/63 | 11.1 |
| FISH del11q | 10/63 | 15.9 |
| FISH del17p | 11/63 | 17.4 |
FISH, fluorescent in situ hybridization.
Mouse cells and tissues
iNKT cells were expanded in vitro for 14 days from total splenocytes of wt mice with 100 ng/mL α-GalCer (Kyowa Hakko Kirin, Tokyo, Japan), 40 U/mL recombinant human interleukin-2 (rhIL-2) (Novartis, Basel, Switzerland), and 10 ng/mL recombinant mouse IL-7 (rmIL-7) (R&D Systems, Minneapolis, MN). To obtain dendritic cells (DCs), bone marrow cells from wt mice were cultured for 5 days with 25 ng/mL granulocyte-macrophage colony-stimulating factor plus 5 ng/mL IL-4 (R&D Systems) and enriched with mouse CD11c beads (Miltenyi Biotec, Bergisch Gladbach, Germany). CLL cells were isolated with a mouse B-cell enrichment kit (EasySep, STEMCELL Technologies) from Tcl1 and Tcl1-CD1d−/− mice. DCs and CLL cells were loaded in vitro with 200 ng/mL α-GalCer. Tcl1 tissues were paraffin included, sectioned, and stained with hematoxylin and eosin.
Human cells
iNKT cells were expanded and purified from healthy donor–derived peripheral blood mononuclear cells (PBMCs) with 100 ng/mL α-GalCer as described.29 CLL cells, healthy donor B cells, and monocytes were purified from PBMCs with a B/CLL isolation kit or anti-CD14 beads (Miltenyi Biotec), respectively. T and CLL cells were depleted with anti-CD3 or anti-CD19 beads, respectively (Miltenyi Biotec).
Flow cytometry
Antibodies were from BioLegend (San Diego, CA) unless otherwise noted. Mouse CLL cells were stained with anti-CD19, anti-immunoglobulin M (anti-IgM), anti-B220, and anti-CD1d monoclonal antibodies (mAbs); mouse iNKT cells were stained with anti-TCRβ mAb and PBS57-loaded mouse CD1d (mCD1d) tetramers (obtained from the National Institutes of Health Tetramer Core Facility). Human cells were analyzed in whole blood: CLL cells were stained with anti-CD5, anti-CD19, and anti-CD1d (BD Biosciences, San Jose, CA) or unlabeled anti-CD1d and goat anti-mouse IgG Abs (Thermo Fisher, Waltham, MA), iNKT cells were stained with anti-CD3 or anti-TCRα/β (BD Biosciences), anti-Vα24, anti-Vβ11 (Coulter, Brea, CA) or anti-Vα24-Jα18 (Miltenyi Biotec), anti-CD4, and anti-CD127 mAbs (Coulter). Human monocytes were stained with anti-CD14 and anti-CD1d. Intracellular cytokine production by iNKT and T cells was determined by using anti-IFN-γ mAbs. Samples were acquired on a FACSCanto II flow cytometer or an LSRFortessa X-20 analyzer (BD Biosciences), excluding dead cells and doublets, and analyzed with FlowJo 10.0.6 software.
iNKT cell activation assays
Murine iNKT cells were activated in vivo by intravenous injection into mouse α-GalCer preloaded DCs, and serum IFN-γ was measured by using enzyme-linked immunosorbent assay (ELISA). Murine iNKT cells were activated in vitro by culturing them with α-GalCer loaded or unloaded CLL cells at a 2:1 effector:target (E:T) ratio with or without 10 μg/mL anti-CD1d blocking mAb (clone 1B1; BioLegend).30 Secreted IFN-γ was tested after 48 hours by ELISA. To activate primary human iNKT cells from CLL patients, PBMCs were depleted from CLL cells and incubated with anti-CD3/anti-CD28 beads (Life Technologies/Invitrogen) (1:1 T cell:bead) or phorbolmyristate acetate (PMA; 25 ng/mL) plus ionomycin (1 μg/mL) (Sigma). After 16 hours, 10 μg/mL Brefeldin A (Sigma) was added for 2 hours before intracellular cytokine staining. To activate healthy iNKT cell lines in vitro, freshly isolated healthy B or CLL cells were cultured at the indicated numbers with 2 × 104 iNKT cells with or without 20 μg/mL blocking anti-CD1d (clone 51.1; BioLegend).31 After 48 hours, secreted IFN-γ was determined by ELISA.
Culture and imaging of NLCs
Frozen lymph node sections were stained with anti-CD68 (Abcam, Ltd., Cambridge, UK) and anti-CD1d (clone 42.1; BD Biosciences) mAbs, followed by labeled goat anti-rabbit and goat anti-mouse Abs (Thermo Fisher), respectively, and 4′,6-diamidino-2-phenylindole (DAPI) (Thermo Fisher) for nuclei counterstaining. NLCs were differentiated in vitro from patient-derived T-cell–depleted PBMCs,6,7 and after 7 to 10 days, they were stained with anti-CD1d mAbs followed by goat anti-mouse Ab heavy and light chains (H&L) (Thermo Fisher) and were fixed and examined by either immunofluorescence or by confocal microscopy upon nuclear counterstaining with DAPI. Purified donor-derived monocytes, allogeneic donor-derived iNKT cells, and purified CD1dneg CLL cells were co-cultured (at a 1:3:5 ratio, respectively) with or without 20 μg/mL anti-CD1d blocking mAb for 7 to 10 days, with 10 U/mL rhIL-2 (Novartis) added only at day 1. T-cell–depleted patient-derived PBMCs were cultured for 14 days with or without allogeneic donor–derived iNKT cells (1:3 iNKT cell to monocyte ratio). Surviving CLL cells were removed from the cultures and counted by Flow-Count Fluorospheres (Coulter). Adherent cells remaining in the cultures were either counted after nuclei labeling with Hoechst stain (Thermo Fisher) or stained with anti-CD68 mAb (BD Biosciences) and DAPI for immunofluorescence and quantitative image analysis. Brightness and contrast were optimized for visualization with ImageJ software.
Statistics
Malignant B-cell expansion kinetics in Tcl1 mice were compared by using nonlinear mixed-effects models32 as reported.21 Differences between groups were assessed by 2-sided (unless otherwise noted) unpaired Student t test or Wilcoxon test for non-Gaussian distributions. Linear correlation between 2 parameter categories was evaluated by Spearman’s test. For multiple group comparisons, 1-way analysis of variance was adopted. A P value < .05 was considered statistically significant. For multivariable analysis of human CLL prognostic factors, a generalized linear model (GLM)33,34 was fitted on the data (see supplemental Data, available on the Blood Web site) using R language and environment for statistical computing.35
Results
iNKT cells control leukemia onset and initial organ infiltration in Tcl1 mice
We investigated the surveillance of CLL cells by iNKT cells in the transgenic leukemia Tcl1 mouse model,25 in which malignant CD19+CD5+IgM+B220low B cells progressively expand from 8 weeks of age (Figure 1A).36 Leukemia cells from Tcl1 mice expressed CD1d at constant levels during disease progression, similar to the normal B-cell counterpart (Figure 1A). Tcl1 mice were crossed with Jα18−/− (Tcl1-Jα18−/−) or with CD1d−/− (Tcl1-CD1d−/−) mice lacking iNKT cells and all CD1d-restricted NK T cells, respectively.17 A longitudinal analysis of CLL peripheral expansion estimated that the leukemia onset and progression were significantly faster in NK T-cell–deficient than in NK T-cell–sufficient Tcl1 mice up to 30 weeks of age (Figure 1B). At later disease stages, the 3 mouse strains exhibited similar rates of CLL progression and by 50 to 55 weeks, they developed full-blown leukemia (Figure 1B) with similar overall survival (data not shown). Histopathology analysis revealed a more aggressive disease infiltration of spleen and lymph nodes in 16- to 17-week-old Tcl1-Jα18−/− mice compared with Tcl1 mice (Figure 1C-D). Conversely, 39- to 43-week-old Tcl1 and Tcl1-Jα18−/− mice exhibited comparable leukemia infiltration scores (Figure 1C-D). To assess the functionality of iNKT cells upon disease progression, we injected α-GalCer preloaded wt DCs in 20- and 50-week-old Tcl1 and wt mice and assessed serum IFN-γ production up to 24 hours later. The cytokine concentration was comparable in 20-week-old Tcl1 and wt mice (Figure 1E). However, at 50 weeks, and irrespective of similar numbers of splenic and liver iNKT cells in leukemic and healthy animals (supplemental Figure 1), IFN-γ production from Tcl1 mice was markedly impaired (Figure 1E), suggesting the acquisition of functional defects by autologous iNKT cells. We then investigated whether normal iNKT cells could directly recognize CD1d-expressing CLL cells in vitro. iNKT cells were directly activated by purified CLL cells from Tcl1 mice but not wt B cells, suggesting the recognition of CD1d-restricted endogenous leukemia-derived lipids. Preloading healthy or leukemia B cells with α-GalCer resulted in robust iNKT cell activation, confirming that both cell types were capable of presenting lipid antigens (Figure 1F). Finally, we investigated whether CD1d expression on CLL cells was a prerequisite for leukemia surveillance by iNKT cells. Purified CLL cells from Tcl1 or Tcl1-CD1d−/− mice were loaded or not loaded with α-GalCer in vitro and then injected into wt animals. The progression of both α-GalCer preloaded CD1d+/+ or CD1d−/− CLL was significantly delayed in blood and spleen compared with unloaded cells (Figure 1G-H), indicating that CD1d expression on CLL was dispensable for leukemia surveillance by iNKT cells.
Lack of NKT cells in Tcl1 mice accelerates leukemia onset and progression. (A) Representative flow cytometry analysis of normal (IgM+B220high) and malignant (IgM+B220low) CD19+ B cells from the blood of the indicated mouse strains at 30 weeks of age and CD1d expression on healthy and malignant B cells in Tcl1 mice at 15 and 45 weeks of age. (B) Longitudinal analysis of leukemia progression in peripheral blood (n = 16 mice per group). Temporal average kinetic curves describing malignant B-cell frequency as a function of age (50-week range) were calculated for each mouse group (thick colored lines) by mixed-effect model analysis. Age of mice at which CLL frequency reaches 50% of the asymptotic value is 28.5 weeks for Tcl1-CD1d−/−, 30.6 weeks for Tcl1-Jα18−/−, and 34.1 weeks for Tcl1 mice. Student t test: Tcl1-CD1d−/− vs Tcl1-Jα18−/− mice, P = .0084; Tcl1-Jα18−/− vs Tcl1 mice, P = .00018. Thin gray curves smoothed via local regression with Gaussian kernel describe the kinetic (30-week) range of malignancy expansion for each animal; the average kinetics (thick colored lines) are superimposed for each strain. (C) CLL disease score of hematoxylin and eosin (H&E)–stained tissue sections from different organs from Tcl1 and Tcl1-Jα18−/− mice at different ages as indicated, defined by a pathologist in a blinded fashion on the basis of the degree of leukemia infiltration and disruption of the normal organ architecture in spleen (SPL), liver (LV), kidney (KD), lung (LG), lymph node (LN), and bone marrow (BM). Box plots depict first quartile, median, and third quartile. The outlier values (circles) less than interquartile range (IQR) –1.5 or greater than IQR +1.5 are shown. Vertical bars represent whiskers indicating the distance from the smallest (lower bar) and the highest (upper bar) nonoutlier values from the first and third quartile, respectively. *P < .05 Wilcoxon test. (D) H&E staining of tissue sections from spleens of 1 representative Tcl1 (left panel) and Tcl1-Jα18−/− (right panel) mouse at 17 weeks of age. The splenic architecture of red pulp (RP) in the Tcl1 mouse is maintained, whereas it is substituted by CLL cells in Tcl1-Jα18−/− mice (original magnification ×40). Images were acquired with Zeiss AxioImager M2m equipped with Nuance FX Multispectral Tissue Imaging System and Nuance Acquisition Software. (E) Tcl1 and wt mice at 20 and 50 weeks of age received α-GalCer–loaded DCs intravenously, and IFN-γ serum level was measured by ELISA at the indicated times. Each curve represents the IFN-γ released by each mouse. One representative result of 3 independent and consistent experiments is shown. **P ≤ .005 Wilcoxon test. (F) Purified CLL from Tcl1 and Tcl1-CD1d−/− mice and purified B cells from wt mice preloaded or not with α-GalCer were cultured for 48 hours with iNKT cell lines from wt mice with or without anti-CD1d blocking mAb as indicated, and the released IFN-γ was measured by ELISA. Graphs depict mean ± standard deviation (SD). One representative result of 2 independent and consistent experiments is shown. *P < .05; **P ≤ .005; ***P ≤ .0005; ****P ≤ .00005 1-way analysis of variance (ANOVA). (G) In all, 2 × 106 purified and α-GalCer preloaded CLL cells from Tcl1 or Tcl1 CD1d−/− mice were injected intraperitoneally into C57BL/6N mice (n = 6 per group). The expansion of circulating CLL cells was monitored by flow cytometry at the indicated time points. One of 2 comparable independent experiments is shown. Graphs depict mean values (symbols) ± SD. *P < .05; **P ≤ .005 Wilcoxon test. (H) Posttransplant spleen infiltration by CLL cells. The number of spleen-infiltrating CD19+IgM+B220low cells from Tcl1 mice was compared 50 days posttransplantation (shown in [G]) with α-GalCer preloaded (α-GalCer) and unloaded (UnL) Tcl1 CLL cells; healthy wt mice were used as control. Bars indicate means ± SD. **P ≤ .005, ***P ≤ .0005; 1-way ANOVA.
Lack of NKT cells in Tcl1 mice accelerates leukemia onset and progression. (A) Representative flow cytometry analysis of normal (IgM+B220high) and malignant (IgM+B220low) CD19+ B cells from the blood of the indicated mouse strains at 30 weeks of age and CD1d expression on healthy and malignant B cells in Tcl1 mice at 15 and 45 weeks of age. (B) Longitudinal analysis of leukemia progression in peripheral blood (n = 16 mice per group). Temporal average kinetic curves describing malignant B-cell frequency as a function of age (50-week range) were calculated for each mouse group (thick colored lines) by mixed-effect model analysis. Age of mice at which CLL frequency reaches 50% of the asymptotic value is 28.5 weeks for Tcl1-CD1d−/−, 30.6 weeks for Tcl1-Jα18−/−, and 34.1 weeks for Tcl1 mice. Student t test: Tcl1-CD1d−/− vs Tcl1-Jα18−/− mice, P = .0084; Tcl1-Jα18−/− vs Tcl1 mice, P = .00018. Thin gray curves smoothed via local regression with Gaussian kernel describe the kinetic (30-week) range of malignancy expansion for each animal; the average kinetics (thick colored lines) are superimposed for each strain. (C) CLL disease score of hematoxylin and eosin (H&E)–stained tissue sections from different organs from Tcl1 and Tcl1-Jα18−/− mice at different ages as indicated, defined by a pathologist in a blinded fashion on the basis of the degree of leukemia infiltration and disruption of the normal organ architecture in spleen (SPL), liver (LV), kidney (KD), lung (LG), lymph node (LN), and bone marrow (BM). Box plots depict first quartile, median, and third quartile. The outlier values (circles) less than interquartile range (IQR) –1.5 or greater than IQR +1.5 are shown. Vertical bars represent whiskers indicating the distance from the smallest (lower bar) and the highest (upper bar) nonoutlier values from the first and third quartile, respectively. *P < .05 Wilcoxon test. (D) H&E staining of tissue sections from spleens of 1 representative Tcl1 (left panel) and Tcl1-Jα18−/− (right panel) mouse at 17 weeks of age. The splenic architecture of red pulp (RP) in the Tcl1 mouse is maintained, whereas it is substituted by CLL cells in Tcl1-Jα18−/− mice (original magnification ×40). Images were acquired with Zeiss AxioImager M2m equipped with Nuance FX Multispectral Tissue Imaging System and Nuance Acquisition Software. (E) Tcl1 and wt mice at 20 and 50 weeks of age received α-GalCer–loaded DCs intravenously, and IFN-γ serum level was measured by ELISA at the indicated times. Each curve represents the IFN-γ released by each mouse. One representative result of 3 independent and consistent experiments is shown. **P ≤ .005 Wilcoxon test. (F) Purified CLL from Tcl1 and Tcl1-CD1d−/− mice and purified B cells from wt mice preloaded or not with α-GalCer were cultured for 48 hours with iNKT cell lines from wt mice with or without anti-CD1d blocking mAb as indicated, and the released IFN-γ was measured by ELISA. Graphs depict mean ± standard deviation (SD). One representative result of 2 independent and consistent experiments is shown. *P < .05; **P ≤ .005; ***P ≤ .0005; ****P ≤ .00005 1-way analysis of variance (ANOVA). (G) In all, 2 × 106 purified and α-GalCer preloaded CLL cells from Tcl1 or Tcl1 CD1d−/− mice were injected intraperitoneally into C57BL/6N mice (n = 6 per group). The expansion of circulating CLL cells was monitored by flow cytometry at the indicated time points. One of 2 comparable independent experiments is shown. Graphs depict mean values (symbols) ± SD. *P < .05; **P ≤ .005 Wilcoxon test. (H) Posttransplant spleen infiltration by CLL cells. The number of spleen-infiltrating CD19+IgM+B220low cells from Tcl1 mice was compared 50 days posttransplantation (shown in [G]) with α-GalCer preloaded (α-GalCer) and unloaded (UnL) Tcl1 CLL cells; healthy wt mice were used as control. Bars indicate means ± SD. **P ≤ .005, ***P ≤ .0005; 1-way ANOVA.
Collectively, these results suggested that iNKT cells controlled the initial CLL expansion phase by mechanisms that may not require direct leukemia cell recognition but became functionally impaired upon disease progression.
High CD1d expression on CLL cells and iNKT cell impairment correlate with disease progression in patients
The above results prompted us to search for a clinical correlate of iNKT cell immune surveillance in a cohort of 68 CLL patients (Table 1). CD1d was expressed on CD19+CD5+ CLL cells with an intrapatient unimodal profile but variable interpatient intensity (Figure 2A). In 46 patients, we determined the circulating iNKT cell frequency (range, 0% to 0.35% of total T cells; median, 0.0235%) and absolute numbers (0 to 0.0056 cells per μL of blood; median, 0.0005 cells per μL of blood). These patients displayed a significant inverse correlation between the level of CD1d expression on CLL cells and the autologous iNKT cell frequencies and counts (Figure 2B). Importantly, CD1d expression on CLL cells was significantly higher and iNKT cell frequency and number were significantly lower in patients with progressive disease than in patients with stable disease (Figure 2Ci,iii,iv). The differential CD1d expression between patients with progressive disease and patients with stable disease was specific for the malignant but not the normal autologous B-cell compartment, which instead expressed comparable (and significantly higher than malignant cells; P < .05) CD1d levels in both stable and progressive cohorts (Figure 2Cii).
High CD1d expression on CLL cells and defective iNKT cells correlate with disease progression in patients. (A) Representative flow cytometry detection of CD1d expression on CD19+CD5+ CLL cells and of iNKT cells in total TCRα/β T cells from 1 patient with stable disease (upper panel) and 1 patient with progressive disease (lower panel). (B) Statistically significant inverse correlation between CD1d expression on CLL cells and autologous iNKT cell frequency (left panel, P = .002551) or iNKT cell number (right panel, P = .005213 Spearman’s correlation test). Scatter plot graphs show single patient value (symbols) with superimposed lines representing the estimated linear regression models (46 patients). (C) Shown are the (i) association of a high CD1d expression on CD19+CD5+ CLL cells (n = 68 patients) with CLL progression, (ii) the absence of significant difference between CD1d expression on nonmalignant CD19+CD5– cells in patients with stable or progressive disease (same cohorts as in [i]), (iii) the correlation between iNKT cell frequency (n = 50 patients), and (iv) number of patients (n = 46) with CLL progression. Box plots depict first quartile, median, third quartile, outliers (circles), and whiskers proportional to IQR. *P < .05; **P < .005; 1-sided Wilcoxon test. (D) Intracellular staining flow cytometry analysis of IFN-γ production by primary circulating iNKT cells from a representative age-matched healthy donor, a CD1dneg/low CLL patient, and a CD1dhigh CLL patient upon ex vivo activation with anti-CD3/anti-CD28 beads or PMA/ionomycin. The percentage of IFN-γ–producing iNKT and T cells is indicated. (E) Frequency of IFN-γ–producing CD4+ (filled circle) and CD4– (empty circle) iNKT cells from age-matched healthy donors, CD1dneg/low CLL patients, and CD1dhigh CLL patients upon α-CD3/α-CD28 bead activation. Graph symbols show single individual values ± SD. *P < .05 Wilcoxon test. (F) Correlation between CD1d expression on CLL cells and the frequency of IFN-γ–producing iNKT cells and T cells upon α-CD3/α-CD28 bead activation. Scatter plot graphs show single patient (n = 15) values for iNKT and T cells (circles) with superimposed lines representing the estimated linear regression models for iNKT cells (solid) and T cells (dashed). Shown is a significant negative correlation between CD1d expression on CLL cells and frequency of IFN-γ–producing iNKT cells (P = .0096) but not T cells (P = .1136; Spearman’s correlation test).
High CD1d expression on CLL cells and defective iNKT cells correlate with disease progression in patients. (A) Representative flow cytometry detection of CD1d expression on CD19+CD5+ CLL cells and of iNKT cells in total TCRα/β T cells from 1 patient with stable disease (upper panel) and 1 patient with progressive disease (lower panel). (B) Statistically significant inverse correlation between CD1d expression on CLL cells and autologous iNKT cell frequency (left panel, P = .002551) or iNKT cell number (right panel, P = .005213 Spearman’s correlation test). Scatter plot graphs show single patient value (symbols) with superimposed lines representing the estimated linear regression models (46 patients). (C) Shown are the (i) association of a high CD1d expression on CD19+CD5+ CLL cells (n = 68 patients) with CLL progression, (ii) the absence of significant difference between CD1d expression on nonmalignant CD19+CD5– cells in patients with stable or progressive disease (same cohorts as in [i]), (iii) the correlation between iNKT cell frequency (n = 50 patients), and (iv) number of patients (n = 46) with CLL progression. Box plots depict first quartile, median, third quartile, outliers (circles), and whiskers proportional to IQR. *P < .05; **P < .005; 1-sided Wilcoxon test. (D) Intracellular staining flow cytometry analysis of IFN-γ production by primary circulating iNKT cells from a representative age-matched healthy donor, a CD1dneg/low CLL patient, and a CD1dhigh CLL patient upon ex vivo activation with anti-CD3/anti-CD28 beads or PMA/ionomycin. The percentage of IFN-γ–producing iNKT and T cells is indicated. (E) Frequency of IFN-γ–producing CD4+ (filled circle) and CD4– (empty circle) iNKT cells from age-matched healthy donors, CD1dneg/low CLL patients, and CD1dhigh CLL patients upon α-CD3/α-CD28 bead activation. Graph symbols show single individual values ± SD. *P < .05 Wilcoxon test. (F) Correlation between CD1d expression on CLL cells and the frequency of IFN-γ–producing iNKT cells and T cells upon α-CD3/α-CD28 bead activation. Scatter plot graphs show single patient (n = 15) values for iNKT and T cells (circles) with superimposed lines representing the estimated linear regression models for iNKT cells (solid) and T cells (dashed). Shown is a significant negative correlation between CD1d expression on CLL cells and frequency of IFN-γ–producing iNKT cells (P = .0096) but not T cells (P = .1136; Spearman’s correlation test).
We next assessed whether the residual circulating iNKT cells in patients with progressive disease were also functionally impaired, as observed in Tcl1 mice with advanced disease. Indeed, iNKT cells from CD1dhigh (relative fluorescence intensity [RFI] ≥3) patients did not respond to activation by either α-CD3/α-CD28 beads or PMA/ionomycin (Figure 2D), whereas autologous T cells displayed selective unresponsiveness only upon activation by α-CD3/α-CD28 beads but not PMA/ionomycin, as reported.37 By contrast, iNKT and T cells were both fully functional in patients with CD1dneg/low (RFI <3) CLL stable disease or healthy donors. Both CD4+ and CD4– iNKT cell subsets38,39 from patients with CD1dhigh CLL produced less IFN-γ and IL-13 upon ex vivo stimulation compared with the cells from patients with CD1dneg/low CLL and healthy age-matched participants (Figure 2D-E; data not shown). IFN-γ production by iNKT cells inversely correlated with CD1d expression (Figure 2F) unlike T cells, suggesting a selective CD1d-related functional impairment of iNKT cells.
Thus, disease progression correlated with CD1d expression by malignant cells and defective iNKT cell counts and functions in CLL patients, akin to that in Tcl1 mice, suggesting an active participation of iNKT cells in leukemia surveillance.
CD1d molecules expressed by CLL cells stimulate normal iNKT cells
We reasoned that the iNKT cell functional and numerical attrition in CD1dhigh CLL patients might result from 2 alternative mechanisms: (1) a persistent CD1d-dependent stimulation by CLL cells, similar to that in T cells during chronic viral infection or malignancies40,41 ; or (2) a defective CD1d-dependent stimulation by CLL cells resulting from dysfunctional antigen-presenting functions of malignant B cells.17,42-44 To address these issues, we cultured iNKT cell lines from healthy donors with purified CD1dneg/low or CD1dhigh CLL cells, or B cells from healthy donors with or without anti-CD1d blocking antibody. IFN-γ was secreted in a CD1d-dependent manner by iNKT cells upon direct recognition of 3 of 4 CD1dhigh but none of 4 CD1dneg/low CLL cells (Figure 3A-B; data not shown). Notably, iNKT cells were more efficiently activated by malignant B cells than by normal B cells, despite similar levels of CD1d expression. The results showed that CLL cells expressing CD1d were able to strongly stimulate iNKT cells in vitro without the addition of α-GalCer, possibly leading to their progressive exhaustion in vivo. Consistent with an exhausted state,41 primary iNKT cells from patients with progressive disease displayed (in addition to the functional unresponsiveness showed above; Figure 2 D-F) a lower IL-7 receptor α chain (CD127) expression compared with cells from patients with stable disease (Figure 3C-D). The reduced CD127 expression was significant for iNKT but not for autologous T cells (Figure 3C-D), further suggesting that iNKT cells might undergo a selective exhaustion process in patients with aggressive CLL.
Differential recognition of CLL and healthy B cells by iNKT cells. (A) CD1d expression by flow cytometry in primary CLL cells and B cells from 4 patients and 4 healthy donors, respectively, used in pairs for iNKT cell activation. (B) The same paired cells were cultured for 48 hours at the indicated numbers with 2 × 104 iNKT cells from healthy donors with or without anti-CD1d (α-CD1d) mAb. IFN-γ release by iNKT cells was measured by ELISA and expressed as mean ± SD. (C) Ex vivo CD127 expression determined by flow cytometry in patients with CD1dneg/low stable CLL vs patients with CD1dhigh progressive CLL. The percentage of CD127-expressing iNKT and T cells is indicated. (D) Frequency of CD127 iNKT and T cells from patients with stable or progressive disease. Graph symbols show single individual values ± SD. ***P < .0005 1-way ANOVA.
Differential recognition of CLL and healthy B cells by iNKT cells. (A) CD1d expression by flow cytometry in primary CLL cells and B cells from 4 patients and 4 healthy donors, respectively, used in pairs for iNKT cell activation. (B) The same paired cells were cultured for 48 hours at the indicated numbers with 2 × 104 iNKT cells from healthy donors with or without anti-CD1d (α-CD1d) mAb. IFN-γ release by iNKT cells was measured by ELISA and expressed as mean ± SD. (C) Ex vivo CD127 expression determined by flow cytometry in patients with CD1dneg/low stable CLL vs patients with CD1dhigh progressive CLL. The percentage of CD127-expressing iNKT and T cells is indicated. (D) Frequency of CD127 iNKT and T cells from patients with stable or progressive disease. Graph symbols show single individual values ± SD. ***P < .0005 1-way ANOVA.
iNKT cells indirectly impair CLL cell survival by restraining proleukemia NLCs in vitro
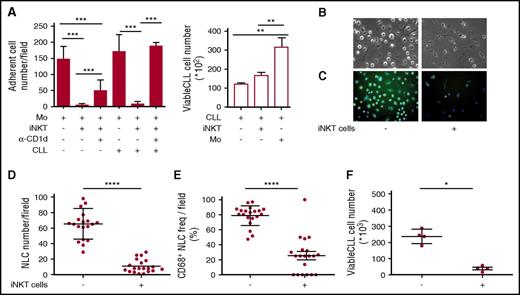
The results obtained above implied that iNKT cells could hinder the progression of CD1d– CLL, raising the question of how this may occur. We hypothesized that iNKT cells might indirectly control leukemia progression by restraining NLCs, which are reported to be CLL-specific TAMs endowed with proleukemia functions.5-7 To address this issue, we first verified the expression of CD1d by NLCs in vivo. Lymph node sections from 2 patients with progressive CLL contained large CD68+ stellate cells that displayed a macrophage-like morphology compatible with that of NLCs that co-expressed CD1d at variable intensity (Figure 4A-B). Immunohistochemistry staining of lymph node sections from a third patient with progressive CLL confirmed the presence of scattered CD1d+ macrophage-like cells surrounded by packed elements that displayed CLL cell morphology (supplemental Figure 2). Irrespective of CD1d expression by autologous CLL cells, CD1d was also expressed by primary monocytes (isolated from patients) that were considered NLC precursors (Figure 4C), as well as by NLCs spontaneously differentiated in vitro upon culturing T-cell–depleted PBMCs from CLL patients (Figure 4D). Second, we investigated the effects of iNKT cells on the differentiation of healthy-donor monocytes into NLCs upon coculture in vitro with purified CLL cells, as described.6,7 CD14+ monocytes were cultured in the presence or absence of allogeneic purified CD1dneg CLL cells and healthy-donor iNKT cells. We used this source of iNKT cells to overcome the quantitative limitations of the clinical samples, assuming that they were functionally comparable with the cells from patients with CD1dneg/low CLL stable disease. After 7 days of culture (Figure 5A), iNKT cells markedly reduced the number of adherent NLCs, which were significantly restored in the presence of anti-CD1d mAbs. Consistent with the absence of CD1d expression on CLL cells, iNKT cells did not directly impair their survival, which was actually sustained by coculture with monocytes (Figure 5A).6
Monocytes and NLCs from CLL patients express CD1d. (A-B) CD1d expression by NLCs in vivo. Frozen lymph node sections from 2 patients with progressive CLL co-stained with anti-CD1d mAb plus goat anti-mouse fluorescein isothiocyanate (FITC) (green), anti-CD68 mAb plus goat anti-rabbit phycoerythrin (PE) (red) Abs, and DAPI (blue). Macrophages co-expressing CD1d and CD68 (yellow arrows) are detectable scattered within (A) dense macrophage infiltrates (original magnification ×400) or in (B) macrophage clusters (original magnification ×200), surrounded by lymphoid elements compatible with CLL cell morphology. Sections were analyzed with a Zeiss AXIO Scope.A1 optical microscope (Zeiss, Oberkochen, Germany). Images were collected with Zeiss Axiocam 503 Color. (C) Representative CD1d expression detected by flow cytometry in circulating monocytes from 1 of 10 CD1dneg CLL patients. (D) CD1d expression by NLCs differentiated in vitro from patients’ PBMCs. Adherent cells were stained with anti-CD1d mAb plus goat anti-mouse PE (green) and DAPI (blue). (Left panel) CD1d expression by immunofluorescence analysis in 1 representative field (original magnification ×20) of 20 to 40 fields per patient performed on 6 patients. (Middle panel) Bright field (gray) merged with CD1d immunofluorescence and DAPI nuclei staining (original magnification ×20) showing NLCs with typical stellate morphology. Immunofluorescence was performed by In Cell Analyzer 1000 GE Healthcare and In Cell Analyzer 1000 Workstation or Arrayscan XTI (Thermo Fisher) with HCS studio software. (Right panel) The same NLCs were stained with anti-CD1d PE mAb and DAPI for confocal imaging. CD1d is localized on the cell surface and in punctate structures (original magnification ×63). Confocal imaging was performed with Confocal Microscope Leica TCS SP2.
Monocytes and NLCs from CLL patients express CD1d. (A-B) CD1d expression by NLCs in vivo. Frozen lymph node sections from 2 patients with progressive CLL co-stained with anti-CD1d mAb plus goat anti-mouse fluorescein isothiocyanate (FITC) (green), anti-CD68 mAb plus goat anti-rabbit phycoerythrin (PE) (red) Abs, and DAPI (blue). Macrophages co-expressing CD1d and CD68 (yellow arrows) are detectable scattered within (A) dense macrophage infiltrates (original magnification ×400) or in (B) macrophage clusters (original magnification ×200), surrounded by lymphoid elements compatible with CLL cell morphology. Sections were analyzed with a Zeiss AXIO Scope.A1 optical microscope (Zeiss, Oberkochen, Germany). Images were collected with Zeiss Axiocam 503 Color. (C) Representative CD1d expression detected by flow cytometry in circulating monocytes from 1 of 10 CD1dneg CLL patients. (D) CD1d expression by NLCs differentiated in vitro from patients’ PBMCs. Adherent cells were stained with anti-CD1d mAb plus goat anti-mouse PE (green) and DAPI (blue). (Left panel) CD1d expression by immunofluorescence analysis in 1 representative field (original magnification ×20) of 20 to 40 fields per patient performed on 6 patients. (Middle panel) Bright field (gray) merged with CD1d immunofluorescence and DAPI nuclei staining (original magnification ×20) showing NLCs with typical stellate morphology. Immunofluorescence was performed by In Cell Analyzer 1000 GE Healthcare and In Cell Analyzer 1000 Workstation or Arrayscan XTI (Thermo Fisher) with HCS studio software. (Right panel) The same NLCs were stained with anti-CD1d PE mAb and DAPI for confocal imaging. CD1d is localized on the cell surface and in punctate structures (original magnification ×63). Confocal imaging was performed with Confocal Microscope Leica TCS SP2.
iNKT cells control CLL survival by restraining proleukemia functions of NLCs. (A) iNKT cells impair NLC generation in vitro. Shown is the number of adherent NLCs differentiated from CD14+ monocytes of healthy donors upon culturing in vitro for 7 days with or without purified CD1dneg CLL cells, healthy iNKT cells, anti-CD1d blocking mAb. (Left panel) Quantitative image analyses of 60 different fields were taken from each well. (Right panel) CLL cells cultured with or without iNKT cells or monocytes were collected and counted by flow cytometry in duplicates. Results are expressed as mean ± SD and are represent 1 of 3 consistent experiments. **P ≤ .005; ***P ≤ .0005 1-way ANOVA. (B-F) iNKT cells impair CLL and NLC survival in vitro. T-cell–depleted PBMCs from a CD1dneg CLL patient were cultured in vitro with or without purified healthy iNKT cells. After 14 days of culture, CLL cells were collected and adherent cells were assessed for morphology in (B) bright field (original magnification ×20) and (C) stained with CD68-FITC (green) and DAPI (blue) for immunofluorescence (original magnification ×20). Quantitative image analyses of 20 different fields were taken directly from the same culture wells to determine (D) total adherent cell counts and (E) CD68+ adherent cell counts. (F) The CLL cells that were harvested from the same wells were counted by flow cytometry in quadruplicate. Bars show mean ± SD. Data refer to 1 of 2 independent experiments performed with different patients that gave comparable results.*P < .05; ****P ≤ .00005 Wilcoxon test. Immunofluorescence and counts were performed by In Cell Analyzer 1000 GE Healthcare and In Cell Analyzer 1000 Workstation or Arrayscan XTI (Thermo Fisher) with HCS studio software. Bright field image acquisitions were performed with Zeiss Axio Observer.z1 with QImaging EXI-Blue equipped with Velocity acquisition software.
iNKT cells control CLL survival by restraining proleukemia functions of NLCs. (A) iNKT cells impair NLC generation in vitro. Shown is the number of adherent NLCs differentiated from CD14+ monocytes of healthy donors upon culturing in vitro for 7 days with or without purified CD1dneg CLL cells, healthy iNKT cells, anti-CD1d blocking mAb. (Left panel) Quantitative image analyses of 60 different fields were taken from each well. (Right panel) CLL cells cultured with or without iNKT cells or monocytes were collected and counted by flow cytometry in duplicates. Results are expressed as mean ± SD and are represent 1 of 3 consistent experiments. **P ≤ .005; ***P ≤ .0005 1-way ANOVA. (B-F) iNKT cells impair CLL and NLC survival in vitro. T-cell–depleted PBMCs from a CD1dneg CLL patient were cultured in vitro with or without purified healthy iNKT cells. After 14 days of culture, CLL cells were collected and adherent cells were assessed for morphology in (B) bright field (original magnification ×20) and (C) stained with CD68-FITC (green) and DAPI (blue) for immunofluorescence (original magnification ×20). Quantitative image analyses of 20 different fields were taken directly from the same culture wells to determine (D) total adherent cell counts and (E) CD68+ adherent cell counts. (F) The CLL cells that were harvested from the same wells were counted by flow cytometry in quadruplicate. Bars show mean ± SD. Data refer to 1 of 2 independent experiments performed with different patients that gave comparable results.*P < .05; ****P ≤ .00005 Wilcoxon test. Immunofluorescence and counts were performed by In Cell Analyzer 1000 GE Healthcare and In Cell Analyzer 1000 Workstation or Arrayscan XTI (Thermo Fisher) with HCS studio software. Bright field image acquisitions were performed with Zeiss Axio Observer.z1 with QImaging EXI-Blue equipped with Velocity acquisition software.
Third, we assessed whether iNKT cells could affect the differentiation of patient-derived NLCs in vitro and, in turn, the survival of CD1dneg CLL cells. T-cell–depleted PBMCs from CD1dneg CLL patients were cultured in the presence or absence of iNKT cell lines from healthy donors. After 14 days of coculture, NLC and CLL counts were markedly reduced in the presence of iNKT cells (Figure 5B,D,F). Furthermore, the adherent cells, differentiated in the absence of iNKT cells, exhibited a macrophage-like shape and homogeneous intracellular CD68 expression typical of NLCs (Figure 5B,C,E). By contrast, the residual adherent cells differentiated in the presence of iNKT cells were mostly spindle shaped and negative for CD68 expression (Figure 5B,C,E). Collectively, these results suggested that iNKT cells can indirectly impair CLL viability by restraining NLC differentiation in a CD1d-depedent manner.
iNKT cell percentage predicts CLL progression in multivariable analysis
In light of the correlation between high CD1d expression by CLL cells and low iNKT cell frequency with disease progression, we compared the prognostic value of these 2 parameters with the currently used markers: Rai clinical stage, ZAP-70 and CD38 expression, chromosomal aberrations, and IGHV gene mutational state of CLL cells. The univariable statistical analysis of our cohort of patients showed that, as expected, disease progression was associated with higher numbers of different chromosomal aberrations, CD38 expression, and unmutated IGHV genes and also with lower iNKT cell frequencies and higher CD1d expression on CLL cells than in stable disease (Table 2; Figure 2C). To predict the progression probability, the above prognostic factors were included as covariates of a GLM in a multivariable analysis. In the GLM-1 model containing all markers, a backward stepwise analysis (supplemental Table 1) selected iNKT cell frequency and CD38 expression on CLL cells as the best significant independent prognostic factors (Spearman correlation test between the 2 covariates, P = .85) (Table 3). A second GLM (GLM-2) (Table 3) was generated on the basis of only the classical CLL prognostic factors, excluding iNKT cell frequency and CD1d expression on CLL cells. By using this procedure, CD38 expression was selected as a unique significant CLL prognostic factor. However, GLM-1 predicted the progression probability of patients with higher sensitivity than GLM-2 (supplemental Table 2), highlighting iNKT cells as a potential novel CLL prognostic parameter.
Univariable analysis defines the prognostic values for CLL of currently used biomarkers, iNKT cells, and CD1d expression
| Prognostic factor . | No. of patients tested . | P* . | Ordering† . |
|---|---|---|---|
| Rai at diagnosis | 60 | .157 | P>S |
| ZAP-70‡ | 59 | .097 | P>S |
| Chromosomal aberration scoreठ| 63 | .034 | P>S |
| FISH del 17p | .243 | P>S | |
| Trisomy 12 | .090 | P>S | |
| FISH del 11q | .083 | P>S | |
| iNKT cell counts‡ | 48 | .050 | P<S |
| CD1d expression‡ | 64 | .032 | P>S |
| IGHV unmutated status | 58 | .003 | P>S |
| iNKT cell frequency‡ | 50 | .003 | P<S |
| CD38‡ | 65 | .0001 | P>S |
| Prognostic factor . | No. of patients tested . | P* . | Ordering† . |
|---|---|---|---|
| Rai at diagnosis | 60 | .157 | P>S |
| ZAP-70‡ | 59 | .097 | P>S |
| Chromosomal aberration scoreठ| 63 | .034 | P>S |
| FISH del 17p | .243 | P>S | |
| Trisomy 12 | .090 | P>S | |
| FISH del 11q | .083 | P>S | |
| iNKT cell counts‡ | 48 | .050 | P<S |
| CD1d expression‡ | 64 | .032 | P>S |
| IGHV unmutated status | 58 | .003 | P>S |
| iNKT cell frequency‡ | 50 | .003 | P<S |
| CD38‡ | 65 | .0001 | P>S |
The distribution of each prognostic factor in patients with stable disease (S) vs progressive disease (P) was compared by Wilcoxon tests.
FISH, fluorescent in situ hybridization.
P values < .05 indicate statistically significant difference between patients with progressive disease and those with stable disease.
P>S tested the hypothesis with prognostic factor values higher among patients with progressive disease than among patients with stable disease; P<S tested the hypothesis with prognostic factor values lower among patients with progressive disease than among patients with stable disease.
Each variable was evaluated as continuous.
Chromosomal aberration score was calculated as the sum of total chromosomal aberration numbers per patient.
Multivariable analysis defines iNKT cell frequency and CD38 expression on CLL as the best factors to predict CLL progression probability
| Model . | Parameter estimate . | SE . | P* . |
|---|---|---|---|
| GLM-1 | |||
| Intercept | 0.23970 | 0.56097 | .6692 |
| iNKT cell frequency† | −8.43884 | 3.33224 | .0113 |
| CD38‡ | 0.02839 | 0.01272 | .0256 |
| GLM-2 | |||
| Intercept | −1.284757 | 0.340107 | .000158 |
| CD38‡ | 0.028270 | 0.009687 | .003519 |
| Model . | Parameter estimate . | SE . | P* . |
|---|---|---|---|
| GLM-1 | |||
| Intercept | 0.23970 | 0.56097 | .6692 |
| iNKT cell frequency† | −8.43884 | 3.33224 | .0113 |
| CD38‡ | 0.02839 | 0.01272 | .0256 |
| GLM-2 | |||
| Intercept | −1.284757 | 0.340107 | .000158 |
| CD38‡ | 0.028270 | 0.009687 | .003519 |
The equation of GLM and the interpretation of parameter and intercept are described in supplemental Data. Best GLM was generated by backward stepwise selection (supplemental Table 1), including all the variables examined by univariable analyses in Table 2. GLM-2 was generated by backward stepwise selection (detailed in supplemental Table 1) by excluding iNKT cell frequency.
SE, standard error.
P < .05 indicates factors with statistically significant prognostic contribution, as calculated by backward stepwise selection, detailed in supplemental Table 1. P values were obtained by means of 2-tailed Z test performed by using R software.
iNKT cell frequency: selected prognostic factor; square root transformation was adopted for iNKT cell frequency to account for the small positive values of iNKT cell frequencies among total T cells.
CD38 expression on CLL is a selected prognostic factor.
Discussion
Our results provide compelling evidence for the involvement of iNKT cells in the immune surveillance against CLL in both mouse models and patients. Indeed, the absence of iNKT cells in Tcl1 mice, a realistic preclinical model of human CLL, accelerated the leukemia onset and initial infiltration (before 30 weeks of age) in secondary lymphoid tissues. Mechanistically, our study suggested that iNKT cells might control CLL cells independent of direct recognition. Moreover, leukemia expanded earlier in Tcl1-CD1d−/− mice that lacked iNKT type I and type II NKT cells compared with Tcl1-Jα18−/− mice that lacked only iNKT cells, which suggests that, at least in CLL, type II NKT cells may also display antitumor functions.17,45 This differs from the immunosuppressive protumor role attributed to type II NKT cells in some solid tumors.17,45 Consistent with previous reports,22,24,46 our data showed CD1d expression on CLL cells from patients, although it was distributed over a wide range of intensity. We found that iNKT cell counts and the expression level of CD1d on CLL cells were inversely correlated. Patients with lower numbers of iNKT cells had higher CD1d expression on CLL cells and were more prone to disease progression, whereas higher numbers of iNKT cell were associated with lower or negative CD1d expression on CLL cells and disease stability. Unexpectedly, lack of CD1d expression by malignant B cells did not represent a mechanism of CLL evasion from iNKT cell surveillance, in sharp contrast with the loss of major histocompatibility complex expression by solid and hematologic tumors escaping from T-cell–dependent selective pressure.47,48
Progressive dysfunctions of iNKT cells were similar in the Tcl1 model and in patients, and both were associated with the maintenance of CD1d expression by CLL cells. The functional defect in Tcl1 mice is intrinsic to iNKT cells because it could not be reversed in vivo by activation with wt DCs preloaded with the potent agonist α-GalCer. Moreover, the impaired IFN-γ production by iNKT cells from patients with CD1dhigh CLL and increased probability of progression is consistent with the functional defect previously described in iNKT cells from cancer patients with poor prognosis.19,20,49 This condition was not described in CLL, possibly because patients were not previously classified depending on either clinical stage (ie, stable or progressive disease) or CD1d expression level on CLL cells.23 Significant immunologic defects have been documented in CLL.42,50-52 In particular, T cells from patients are hyporesponsive to α-CD3/CD28 triggering, whereas they produce more cytokines than healthy T cells upon PMA/ionomycin activation.37,53 By contrast, iNKT cells from patients with progressive CLL disease did not respond to either anti-CD3/CD28 or to PMA/ionomycin activation, suggesting the acquisition of distinct, more profound defects compared with T cells. Moreover, iNKT cell, but not T-cell, responsiveness inversely correlated with CD1d levels on CLL cells, indicating that iNKT cell dysfunctions might derive from a CD1d-cognate contact with CLL cells. In this respect, we indeed showed that in the absence of α-GalCer, primary CLL cells from Tcl1 mice, but not wt B cells, stimulated iNKT cells in vitro in a CD1d-cognate manner. Furthermore, primary human CD1dhigh CLL cells, but not healthy B cells, potently activated iNKT cells without the addition of α-GalCer. These findings suggest that iNKT cells can directly recognize leukemia-derived lipids presented by CD1d on CLL cells. As a result of this, our findings also support the hypothesis that iNKT cells might become functionally exhausted upon chronic stimulation by CD1d-expressing CLL cells. We indeed found a reduced CD127 expression by iNKT cells in CD1dhigh progressive CLL, similar to the phenotype acquired by exhausted T cells in the course of chronic viral infections.41 Because IL-7 signaling via CD127 is critical for iNKT cell homeostasis,54,55 its decline might account for the observed loss of these cells upon CLL progression. Nevertheless, additional factors, such as immunosuppression or impaired costimulation delivered by CLL cells9 may also contribute to iNKT cell hyporesponsiveness in patients with progressive disease, similar to T-cell exhaustion in CLL patients.37
We faced the intriguing situation in which an intact iNKT cell response correlated with low or absent CD1d expression by CLL cells in patients with stable disease. This suggested that iNKT cells might also hinder CLL progression independently of CD1d cognate recognition of leukemia cells. It has been demonstrated that iNKT cells can indirectly control neuroblastoma or melanoma via CD1d-dependent killing of TAMs16 or modulation of suppressive myeloid cells,56 respectively. Indeed, we show that α-GalCer preloaded mouse CLL cells that do or do not express CD1d were equally controlled when injected into wt mice, suggesting that CD1d expression by leukemia cells is dispensable for iNKT cell control. Moreover, we provide evidence that human iNKT cells can control CLL survival indirectly by restraining in vitro proleukemia functions of NLCs, a CLL-specific TAM population5,8 capable of supporting leukemia within protective niches of lymphoid tissues. Indeed, we identified CD68+ macrophage-like NLCs expressing CD1d in CLL lymph nodes from patients with progressive disease. Our results are relevant in light of recent findings that support a critical pathogenic role for macrophages in CLL. For instance, loss of macrophage migration inhibitory factor, responsible for NLC recruitment, delays CLL development in TCL1-tg/Mif−/− mice,57 whereas the therapeutic targeting of NLCs leads to decreased leukemia progression in preclinical models of mouse and human CLL.11
In line with the biological results supporting the role of iNKT cells in CLL control, our multivariable analysis identified iNKT cell frequency as an independent parameter that significantly predicts disease progression in association with CD38 expression. Further validation of the prognostic power of iNKT cell frequency is needed in an independent cohort of CLL patients.
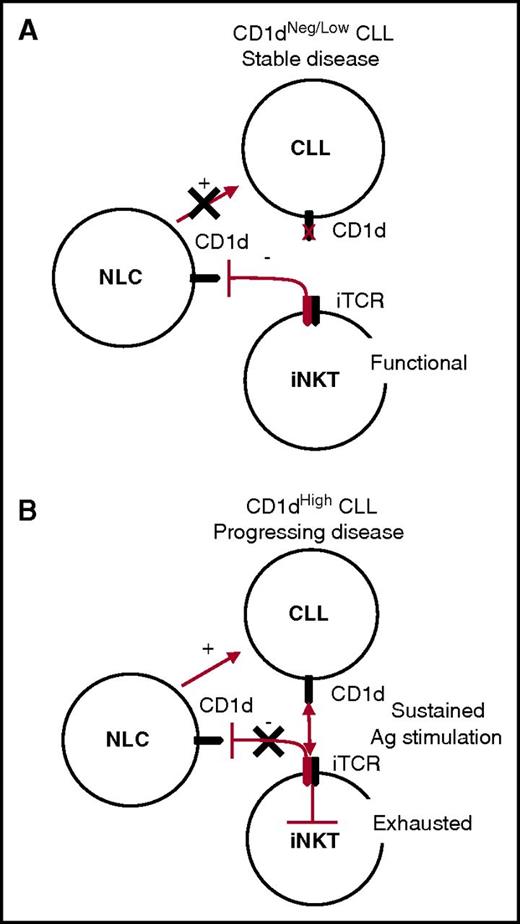
On the basis of the above considerations, we can envisage a unifying model for the iNKT cell immune surveillance of CLL, whereby the control of NLCs remains active in the long term in patients with stable CD1dneg/low CLL, indirectly hindering leukemia progression. By contrast, iNKT cell immune surveillance becomes impaired in patients with progressive CD1dhigh CLL, owing to the persistent antigen stimulation of iNKT cells that ultimately leads to their functional exhaustion and has an impact on their NLC control (Figure 6).
Unifying model for CLL surveillance by iNKT cells. (A) Stable disease correlates with absent or low CD1d CLL expression by CLL cells. In this situation, iNKT cells maintain normal frequencies and functions and the capability to constrain NLCs through a CD1d-cognate interaction, which has a negative impact on CLL survival. (B) Progressing disease correlates with high CD1d expression by CLL cells. In this situation, iNKT cells are persistently hyperstimulated by CLL cells in an antigen (Ag)-dependent fashion, leading to their exhaustion. As a result, iNKT cells no longer control NLCs, which can provide unconstrained support for CLL cell survival and proliferation.
Unifying model for CLL surveillance by iNKT cells. (A) Stable disease correlates with absent or low CD1d CLL expression by CLL cells. In this situation, iNKT cells maintain normal frequencies and functions and the capability to constrain NLCs through a CD1d-cognate interaction, which has a negative impact on CLL survival. (B) Progressing disease correlates with high CD1d expression by CLL cells. In this situation, iNKT cells are persistently hyperstimulated by CLL cells in an antigen (Ag)-dependent fashion, leading to their exhaustion. As a result, iNKT cells no longer control NLCs, which can provide unconstrained support for CLL cell survival and proliferation.
In conclusion, this study supports a role for the iNKT cell/CD1d axis in the control of CLL and highlights iNKT cells as new possible prognostic markers for disease progression.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Maurilio Ponzoni (Pathology Unit, San Raffaele Scientific Institute, Milan, Italy), for blindly scoring CLL infiltration in Tcl1 mice and Claudio Tripodo (Tumor Immunology Unit, Department of Health Science, Human Pathology Section, University of Palermo, Palermo, Italy), for insightful discussions on human CLL tissue evaluation. The authors also thank the National Institutes of Health Tetramer Core Facility for providing the mouse PBS57-loaded mCD1d-PE tetramers. Imaging analysis was performed with technical support and equipment from the Advanced Light and Electron Microscopy Bioimaging Center, San Raffaele Scientific Institute, Milan, Italy.
This study was supported by the Italian Association for Cancer Research Program on Molecular Clinical Oncology, grant 5 per mille n.9965.
F.G. is a PhD candidate in the International PhD Course of Molecular Medicine, Università Vita-Salute San Raffaele, and this work is submitted in fulfillment of the requirement for a PhD.
Authorship
Contribution: C.d.L. designed and performed the experiments and analyzed data regarding CLL patients; F.G. designed and performed the experiments and analyzed the data generated in murine models; L.A. performed statistical analyses; G.D. contributed to experiments in murine models; L.S. and C.S. managed patients, the clinical database, and CLL sample collection; P.R. provided PBMC and CLL cells isolated from patient blood samples; M.T.B. contributed to Tcl1 mouse maintenance; A.G. performed analysis of human CLL lymph node sections; C.D. discussed the blind scoring of murine tissues; A.D.N. provided human CLL lymph nodes; M.C. and C.F. provided human blood samples from age-matched healthy donors; C.S., P.G., F.C.-C., and M.B. discussed results and manuscript writing; and P.D., G.C., and C.d.L. conceived the study and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Paolo Dellabona, Division of Immunology, Transplantation and Infectious Diseases, San Raffaele Scientific Institute, Via Olgettina 58, 20132 Milan, Italy; e-mail: dellabona.paolo@hsr.it; Giulia Casorati, Division of Immunology, Transplantation and Infectious Diseases, San Raffaele Scientific Institute, Via Olgettina 58, 20132 Milan, Italy; e-mail: casorati.giulia@hsr.it; and Claudia de Lalla, Division of Immunology, Transplantation and Infectious Diseases, San Raffaele Scientific Institute, Via Olgettina 58, 20132 Milan, Italy; e-mail: delalla.claudia@hsr.it.
References
Author notes
P.D., G.C., and C.d.L. contributed equally to this study as senior authors.

![Figure 1. Lack of NKT cells in Tcl1 mice accelerates leukemia onset and progression. (A) Representative flow cytometry analysis of normal (IgM+B220high) and malignant (IgM+B220low) CD19+ B cells from the blood of the indicated mouse strains at 30 weeks of age and CD1d expression on healthy and malignant B cells in Tcl1 mice at 15 and 45 weeks of age. (B) Longitudinal analysis of leukemia progression in peripheral blood (n = 16 mice per group). Temporal average kinetic curves describing malignant B-cell frequency as a function of age (50-week range) were calculated for each mouse group (thick colored lines) by mixed-effect model analysis. Age of mice at which CLL frequency reaches 50% of the asymptotic value is 28.5 weeks for Tcl1-CD1d−/−, 30.6 weeks for Tcl1-Jα18−/−, and 34.1 weeks for Tcl1 mice. Student t test: Tcl1-CD1d−/− vs Tcl1-Jα18−/− mice, P = .0084; Tcl1-Jα18−/− vs Tcl1 mice, P = .00018. Thin gray curves smoothed via local regression with Gaussian kernel describe the kinetic (30-week) range of malignancy expansion for each animal; the average kinetics (thick colored lines) are superimposed for each strain. (C) CLL disease score of hematoxylin and eosin (H&E)–stained tissue sections from different organs from Tcl1 and Tcl1-Jα18−/− mice at different ages as indicated, defined by a pathologist in a blinded fashion on the basis of the degree of leukemia infiltration and disruption of the normal organ architecture in spleen (SPL), liver (LV), kidney (KD), lung (LG), lymph node (LN), and bone marrow (BM). Box plots depict first quartile, median, and third quartile. The outlier values (circles) less than interquartile range (IQR) –1.5 or greater than IQR +1.5 are shown. Vertical bars represent whiskers indicating the distance from the smallest (lower bar) and the highest (upper bar) nonoutlier values from the first and third quartile, respectively. *P < .05 Wilcoxon test. (D) H&E staining of tissue sections from spleens of 1 representative Tcl1 (left panel) and Tcl1-Jα18−/− (right panel) mouse at 17 weeks of age. The splenic architecture of red pulp (RP) in the Tcl1 mouse is maintained, whereas it is substituted by CLL cells in Tcl1-Jα18−/− mice (original magnification ×40). Images were acquired with Zeiss AxioImager M2m equipped with Nuance FX Multispectral Tissue Imaging System and Nuance Acquisition Software. (E) Tcl1 and wt mice at 20 and 50 weeks of age received α-GalCer–loaded DCs intravenously, and IFN-γ serum level was measured by ELISA at the indicated times. Each curve represents the IFN-γ released by each mouse. One representative result of 3 independent and consistent experiments is shown. **P ≤ .005 Wilcoxon test. (F) Purified CLL from Tcl1 and Tcl1-CD1d−/− mice and purified B cells from wt mice preloaded or not with α-GalCer were cultured for 48 hours with iNKT cell lines from wt mice with or without anti-CD1d blocking mAb as indicated, and the released IFN-γ was measured by ELISA. Graphs depict mean ± standard deviation (SD). One representative result of 2 independent and consistent experiments is shown. *P < .05; **P ≤ .005; ***P ≤ .0005; ****P ≤ .00005 1-way analysis of variance (ANOVA). (G) In all, 2 × 106 purified and α-GalCer preloaded CLL cells from Tcl1 or Tcl1 CD1d−/− mice were injected intraperitoneally into C57BL/6N mice (n = 6 per group). The expansion of circulating CLL cells was monitored by flow cytometry at the indicated time points. One of 2 comparable independent experiments is shown. Graphs depict mean values (symbols) ± SD. *P < .05; **P ≤ .005 Wilcoxon test. (H) Posttransplant spleen infiltration by CLL cells. The number of spleen-infiltrating CD19+IgM+B220low cells from Tcl1 mice was compared 50 days posttransplantation (shown in [G]) with α-GalCer preloaded (α-GalCer) and unloaded (UnL) Tcl1 CLL cells; healthy wt mice were used as control. Bars indicate means ± SD. **P ≤ .005, ***P ≤ .0005; 1-way ANOVA.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/129/26/10.1182_blood-2016-11-751065/4/m_blood751065f1.jpeg?Expires=1767841277&Signature=SViPKRVlL~cvFHVr7l0gMakzgS1h~JbwgE0B8j1XDQdlB-o8CDyoPGlTOkZNeySOg-aKTZbts00loyhNUibT9VACYj6Gs3a4EPyWhnrjcXoU9bbFv52~u5zCoRVn1NgG4~PlvDcUKYiADT2Mdtjc4ZM5Rwmj~qaQM4qnojMbTYvBvRQq2MXz7bJULjke~4AmQ-YqfyfR40h0av2PkL~u8fb0GI9UjYWaE8OlJaNmO~ua3eSUqhY6RNMesd4i~JpiWAPmcMwVdX1xyd2HIhW1p5AZ-qBW2zZlSDGc7ERTwziVhcnmzthMZNmvciUhbSaez67B4Zo2l1QwlmH73JC1Ew__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 2. High CD1d expression on CLL cells and defective iNKT cells correlate with disease progression in patients. (A) Representative flow cytometry detection of CD1d expression on CD19+CD5+ CLL cells and of iNKT cells in total TCRα/β T cells from 1 patient with stable disease (upper panel) and 1 patient with progressive disease (lower panel). (B) Statistically significant inverse correlation between CD1d expression on CLL cells and autologous iNKT cell frequency (left panel, P = .002551) or iNKT cell number (right panel, P = .005213 Spearman’s correlation test). Scatter plot graphs show single patient value (symbols) with superimposed lines representing the estimated linear regression models (46 patients). (C) Shown are the (i) association of a high CD1d expression on CD19+CD5+ CLL cells (n = 68 patients) with CLL progression, (ii) the absence of significant difference between CD1d expression on nonmalignant CD19+CD5– cells in patients with stable or progressive disease (same cohorts as in [i]), (iii) the correlation between iNKT cell frequency (n = 50 patients), and (iv) number of patients (n = 46) with CLL progression. Box plots depict first quartile, median, third quartile, outliers (circles), and whiskers proportional to IQR. *P < .05; **P < .005; 1-sided Wilcoxon test. (D) Intracellular staining flow cytometry analysis of IFN-γ production by primary circulating iNKT cells from a representative age-matched healthy donor, a CD1dneg/low CLL patient, and a CD1dhigh CLL patient upon ex vivo activation with anti-CD3/anti-CD28 beads or PMA/ionomycin. The percentage of IFN-γ–producing iNKT and T cells is indicated. (E) Frequency of IFN-γ–producing CD4+ (filled circle) and CD4– (empty circle) iNKT cells from age-matched healthy donors, CD1dneg/low CLL patients, and CD1dhigh CLL patients upon α-CD3/α-CD28 bead activation. Graph symbols show single individual values ± SD. *P < .05 Wilcoxon test. (F) Correlation between CD1d expression on CLL cells and the frequency of IFN-γ–producing iNKT cells and T cells upon α-CD3/α-CD28 bead activation. Scatter plot graphs show single patient (n = 15) values for iNKT and T cells (circles) with superimposed lines representing the estimated linear regression models for iNKT cells (solid) and T cells (dashed). Shown is a significant negative correlation between CD1d expression on CLL cells and frequency of IFN-γ–producing iNKT cells (P = .0096) but not T cells (P = .1136; Spearman’s correlation test).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/129/26/10.1182_blood-2016-11-751065/4/m_blood751065f2.jpeg?Expires=1767841277&Signature=uk0O3o6Un4jiFw982XUUeJVBL6f1dJmOzwMp-2DBMXlbzGORNUcvix87EEGMUCiNl7XJNz5Us4eO-L23q8MzqbKCrxnmTLFwiJPNrGAtEyE~A8wHyeCZY~nRFkNhZZ0WQE52GZD1pyvnStAWKsRbOznlUGexAQc46RllYxWzwEIUaim9cIhKN7HxlcAVNJgeswRAGUgaGC59tzRGqok5S-Vnaa30BtT1b7zgvfq8kmnqPU4gR~ORPa6gZsRFHmENsKzkqKfNkA94k09rrIGUmONmORP~~S-PLKmLACYy-snHd13pqi9tQC~1RAbxUceKgPFBeQw-uMNgGR775PRjjQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)