Key Points
Next-generation functional genomics identifies B-cell development genes, pathways, and feedback loops that affect dex activity in B-ALL.
Suppression of lymphoid-restricted PI3Kδ synergizes with dex in B-ALL by enhancing or restoring regulation of cell-death genes.
Abstract
Glucocorticoids (GCs), including dexamethasone (dex), are a central component of combination chemotherapy for childhood B-cell precursor acute lymphoblastic leukemia (B-ALL). GCs work by activating the GC receptor (GR), a ligand-induced transcription factor, which in turn regulates genes that induce leukemic cell death. Which GR-regulated genes are required for GC cytotoxicity, which pathways affect their regulation, and how resistance arises are not well understood. Here, we systematically integrate the transcriptional response of B-ALL to GCs with a next-generation short hairpin RNA screen to identify GC-regulated “effector” genes that contribute to cell death, as well as genes that affect the sensitivity of B-ALL cells to dex. This analysis reveals a pervasive role for GCs in suppression of B-cell development genes that is linked to therapeutic response. Inhibition of phosphatidylinositol 3-kinase δ (PI3Kδ), a linchpin in the pre-B-cell receptor and interleukin 7 receptor signaling pathways critical to B-cell development (with CAL-101 [idelalisib]), interrupts a double-negative feedback loop, enhancing GC-regulated transcription to synergistically kill even highly resistant B-ALL with diverse genetic backgrounds. This work not only identifies numerous opportunities for enhanced lymphoid-specific combination chemotherapies that have the potential to overcome treatment resistance, but is also a valuable resource for understanding GC biology and the mechanistic details of GR-regulated transcription.
Introduction
Although glucocorticoids (GCs) have been used to treat lymphoid malignancies for over half a century,1 the mechanism of their cytotoxicity is still not clear. Nonetheless, GC-based combination chemotherapy protocols are effective, particularly in children with B-cell precursor acute lymphoblastic leukemia (B-ALL). Although ∼90% of children on these protocols are cured, there are few effective treatments for the 10% who do not respond to this therapy.1 Importantly, response to GCs alone is a good predictor of overall response to chemotherapy, indicating a central role for GCs in overall treatment efficacy and suggesting that the outcomes for resistant patients may be improved by enhancing GC potency.1 Unfortunately, simply enhancing GC potency runs the risk of proportional increases in debilitating side effects, such as avascular necrosis and diabetes mellitus. The goal of this work is to determine how GCs kill B-ALL and then systematically identify targets that enhance the lymphoid-specific potency of GCs in resistant patients.
GCs, such as dexamethasone (dex), induce cell death through the GC receptor (GR), a ligand-activated transcription factor whose transcriptional activity is required for GC cytotoxicity.1 GR regulates gene expression by binding DNA and nucleating the assembly of regulatory cofactors. Mutations in specific GR cofactors (CREBBP,2 NCOR1, and TBL1XR12,3 ) disrupt GC-induced gene regulation in B-ALL and have been associated with GC resistance. Dozens of GR-regulated genes have also been correlated with efficacy in B-ALL. Most prominently, repression of antiapoptotic BCL2 and simultaneous activation of proapoptotic BIM (BCL2L11) have been shown to tip the apoptotic balance of B-ALL toward cell death.1 Regulation of these genes may be direct but also involves a feed-forward loop with KLF13, disruption of which results in high BCL2 expression and resistance.1 GCs also increase expression of thioredoxin-interacting protein (TXNIP), which induces cell death by increasing reactive oxygen species and/or blocking glucose transport, effectively starving cells.1 Other studies have shown that GCs may induce cell death by increasing glycolysis (via PFKFB2, PGK1, and PFKP1 ),4 exhausting the depleted glycolytic reserves of lymphoid cells. Taken together, these studies suggest that dex-induced cell death is multifactorial, with faithful GR-driven gene regulation being essential for overall treatment response.
About 2 dozen genetic lesions have been associated with overall treatment resistance or relapse in B-ALL. In addition to mutation of GR cofactors, larger chromosomal changes such as hypodiploidy, t(9;22) (BCR-ABL), t(4;11) (MLL-AF4), and PR2Y8-CRLF2 have been associated with poor prognosis,1 but not resistance, to a specific chemotherapeutic agent. Furthermore, a growing number of resistance-associated lesions have been identified in factors that are involved in B-cell development, including CDKN2A/B, RAS, IZKF1, VREB1, and PAX5, but have not been mechanistically linked to treatment failure.2 Thus, how the majority of these lesions affect treatment response is not known.
Methods
Cell lines, patient specimens, and reagents
B-ALL cell lines (697, B1, KASUMI-2, KOPN-8, MHH-CALL4, MUTZ5, NALM-6, RCH-ACV, RS4;11, and SUP-B15) were obtained from Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ) or ATCC, who validated their genetic background, and then screened for mycoplasma contamination. The background of patient samples (HM2872, HM3101, HM3722) was tested by the Children's Oncology Group. The patient-derived xenograft (ALL121) was genetically characterized previously.1 B-cell lines and specimens were grown and maintained at 37°C, 5% CO2, in RPMI 1640 medium ((+)L-Glutamine) supplemented with 10% fetal bovine serum, unless otherwise noted. HEK293T cells (Clontech) were grown under the same conditions in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum on polylysine-coated plates. Cells were treated with dex (Sigma-Aldrich) or CAL-101 (Selleck Chemicals) dissolved in ethanol.
Gene expression microarrays
Illumina HT12 v4 microarrays were used to measure differential regulation of gene expression by dex. Cell lines and patient specimens were treated with 1 µM dex or ethanol control for 4 hours. RNA was isolated (Qiagen miRNAeasy) and run on arrays at the University of California, Los Angeles (UCLA) Neurosciences Genomics Core (UNGC). At least 3 biological repeats were performed for each sample. Arrays were processed using the R/Bioconductor lumi package.1 Batch effects were corrected using Combat from the SVA package.2 Differential expression of dex-treated vs vehicle was then calculated using ebayes from the limma package.1 False discovery rate was calculated using Benjamini-Hochberg and q value (qvalue package), each producing similar results. Data are available from the Gene Expression Omnibus (GEO; GSE94302).
Differential expression analysis
We used previously published xenograft data5 to validate and lend power to dex-regulated genes identified in our laboratory (GEO no. GSE57795). We processed these arrays as described in the previous section, then combined the results with our data and filtered. A 2-sided Kolmogorov-Smirnov (KS) test was used to determine which genes were persistently upregulated or downregulated across all samples using a q value of 10−4.
Clustering of regulated genes based on differential expression was performed using Euclidean distance in R. Principal component analysis was used on differentially regulated genes to determine the similarity of response to treatment; Ingenuity Pathway Analysis software (Qiagen) was used to perform pathway and gene ontology analysis of differentially regulated genes.
Additional methods
Chromatin immunoprecipitation followed by deep sequencing (ChIP-seq),1 viral preparation,1 short hairpin RNA (shRNA) screening,2 cloning of individual shRNAs and knockdown,6 quantitative polymerase chain reaction,7 western blotting,8 cell viability,9 and patient-derived xenograft models10,11 were performed largely as previously described with additional details provided in supplemental Methods (available on the Blood Web site).
Results
Dex regulates B-cell development genes
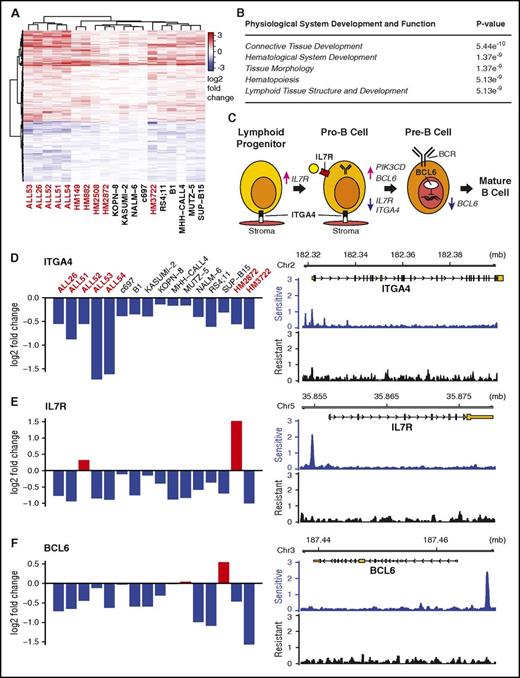
We integrated 2 complementary technologies to determine how GCs induce cell death in B-ALL: dex-induced differential gene expression analysis and functional genomics by large-scale shRNA gene knockdown. By combining these methods, we identified “effector genes”: those GR-regulated genes that drive glucocorticoid-induced cell death in B-ALL. We first isolated the primary effects of GCs in sensitive B-ALL samples by measuring immediate (4-8 hour) changes in gene expression in response to high-dose dex. Using 19 human B-ALL cell lines, primary patient specimens, and existing data from patient-derived xenograft models (PDXs),12 we found that only 4 genes were significantly regulated (q ≤ 0.05) in each sample: FKBP5, TSC22D3 (GiLZ), SMAP2, and TXNIP. Of these, only TXNIP and GiLZ have been previously linked to dex-induced cell death.13 However, we identified another 588 genes that are consistently activated or repressed across samples (KS test, adjusted P value ≤ 1e-4), which we term commonly regulated genes (CRGs) (Figure 1A; supplemental Document 2). Consistent with previous studies, CRGs include BCL2, BCL2L11, KLF13, ZBTB16, and GR itself.14,15 Pathway and gene ontology analyses not only identified expected general GC functions, including diabetes and cell death and survival (supplemental Figure 1A-B), but also a previously unobserved enrichment for hematological and lymphoid development (Figure 1B). Within this category, dex repressed expression of 3 genes, ITGA4,16 IL7R,14 and BCL6,17 which are key factors in early B-cell development (Figure 1C). Dex also repressed genes related to B-cell receptor (BCR) signaling (CD79B, CSK, FYN, BTK, PIK3CD, PIK3C2B, PIK3R2) and activated CXCR4, a receptor that homes B cells to the bone marrow and germinal centers for further maturation.18 These regulatory patterns suggest a pervasive role for GCs in B-cell development and provide a mechanistic explanation for observations made 30 years ago that GCs have a negative effect on early B-cell development.19,20
Dex regulates B-cell development genes in sensitive B-ALL samples. (A) Heatmap clustering genes commonly regulated (KS test, q ≤ 10−4) by dex across 16 samples. Primary and PDX samples are marked red; cell lines, black. (B) Ingenuity pathway analysis of regulated genes shows enrichment for hematological development genes. (C) Stop or push through model for dex in B-cell development highlighting the roles of dex-repressed ITGA4, IL7R, and BCL6. (D-F) Differential gene expression values across sensitive B-ALL sample across samples measured by microarray (left) and GR occupancy in sensitive (B1) and resistant (HM3101) samples measured by ChIP-seq in response to dex suggest ITGA4, IL7R, and BCL6 are direct targets of GR regulation.
Dex regulates B-cell development genes in sensitive B-ALL samples. (A) Heatmap clustering genes commonly regulated (KS test, q ≤ 10−4) by dex across 16 samples. Primary and PDX samples are marked red; cell lines, black. (B) Ingenuity pathway analysis of regulated genes shows enrichment for hematological development genes. (C) Stop or push through model for dex in B-cell development highlighting the roles of dex-repressed ITGA4, IL7R, and BCL6. (D-F) Differential gene expression values across sensitive B-ALL sample across samples measured by microarray (left) and GR occupancy in sensitive (B1) and resistant (HM3101) samples measured by ChIP-seq in response to dex suggest ITGA4, IL7R, and BCL6 are direct targets of GR regulation.
To identify which CRGs are likely direct targets of GR, we performed ChIP-seq for GR in GC-sensitive and -resistant cells. In the GC-sensitive human ALL cell line B1, we observed a greater number of stronger GR-binding sites (∼50 000 sites) compared with a relapsed primary human B-ALL specimen, HM3101 (∼30 000 sites). Although <3% of the binding sites overlapped between samples, a significant fraction were within 10 kb of CRGs (supplemental Figure 2A-B). Importantly, binding sites were enriched near CRGs in sensitive compared with resistant cells (supplemental Figure 2C-D), including ITGA4, IL7R, and BCL6, suggesting that they are direct targets of GR (Figure 1D-F). The striking shift in binding pattern suggests a difference in the accessibility of binding sites (as in John et al21 ), the viability of transcriptional cofactors that direct GR binding (as in Chodankar et al22 ), or a previously unrecognized signaling pathway that changes GR function. Most importantly, this demonstrates that GR can be active in resistant B-ALL, but also fail to bind and regulate effector genes.
Next-generation shRNA screen identifies effectors of GC-induced cell death
Next, we conducted a large-scale next-generation RNA interference screen to determine which GR-regulated genes contribute to cell death and to pinpoint pathways that modulate GC potency.11,23 We identified an appropriate cell line model for screening by comparing the dex-induced transcriptional response of cell lines to primary specimens. Although the messenger RNA levels in unstimulated specimens were different (data not shown), the dex-induced changes in cell lines were similar to the patient specimens and PDX samples (Figure 1; supplemental Figure 1C-D), indicating that cell lines are a reasonable model for dex response in B-ALL. We chose NALM-6 cells, which have intermediate dex sensitivity and relatively rapid growth.
The next-generation screen is composed of an ultra-complex shRNA library10,11,23 that has 4 advantages over other screens: (1) thousands of negative control shRNAs are included to increase statistical confidence and identification of true hits; (2) the large number (25) of shRNAs per gene decreases both the false-positive and false-negative rates; (3) the shRNAs are more active,24 allowing a quantitative analysis of gene knockdown; and (4) both synthetic interactions (the effect of knockdown on dex sensitivity) and the effect of knockdown on NALM-6 growth can be calculated from the data. We adapted how the screen was performed previously11 (see “Methods”) using an intermediate dose of dex (35 nM) to enable identification of sensitizing and protective hits. The 4 most relevant of 11 total panels were screened (cancer, apoptosis, gene expression, kinases [CAGEK]), comprising ∼5800 genes and >140 000 shRNAs, covering about 40% of the CRGs (Figure 2A; supplemental Figure 3A).
Next-generation shRNA screen identifies sources of sensitivity and resistance to dex in B-ALL. (A) Venn diagrams showing that 247 of the CRGs are covered by the screen, 63 of which affect dex sensitivity. (B) Volcano plot of the effect of shRNA gene knockdown on dex sensitivity. Each point is a gene with the log significance on the y-axis, with relative effect (phenotype) on dex-induced cell death on the x-axis. GR is the most protective when knocked down, and knockdown of PIK3CD makes NALM-6 cell more sensitive. Top hits (Mann-Whitney, P ≤ .05) are green: sensitizing; purple: protective; gray: P > .05. (C) Zoom of volcano plot showing genes commonly mutated in treatment resistant or relapsed patients with B-ALL have an effect on dex sensitivity when knocked down (supplemental Table 1). (D) Identification of effector genes from among the CRGs. Plot of dex sensitivity phenotype when knocked down (x-axis) vs the average change in expression in response to dex (y-axis) for genes that are significantly regulated by dex and are top hits in the screen. Genes validated as effectors of dex-induced cell death are either: (1) downregulated by dex and cause sensitivity when knocked down (green shaded) or (2) upregulated by dex and are protective when knocked down (purple shaded). Genes involved in B-cell development or previously identified as effectors are in bold.
Next-generation shRNA screen identifies sources of sensitivity and resistance to dex in B-ALL. (A) Venn diagrams showing that 247 of the CRGs are covered by the screen, 63 of which affect dex sensitivity. (B) Volcano plot of the effect of shRNA gene knockdown on dex sensitivity. Each point is a gene with the log significance on the y-axis, with relative effect (phenotype) on dex-induced cell death on the x-axis. GR is the most protective when knocked down, and knockdown of PIK3CD makes NALM-6 cell more sensitive. Top hits (Mann-Whitney, P ≤ .05) are green: sensitizing; purple: protective; gray: P > .05. (C) Zoom of volcano plot showing genes commonly mutated in treatment resistant or relapsed patients with B-ALL have an effect on dex sensitivity when knocked down (supplemental Table 1). (D) Identification of effector genes from among the CRGs. Plot of dex sensitivity phenotype when knocked down (x-axis) vs the average change in expression in response to dex (y-axis) for genes that are significantly regulated by dex and are top hits in the screen. Genes validated as effectors of dex-induced cell death are either: (1) downregulated by dex and cause sensitivity when knocked down (green shaded) or (2) upregulated by dex and are protective when knocked down (purple shaded). Genes involved in B-cell development or previously identified as effectors are in bold.
The data from the screen were robust, sensitive, and consistent with the known features of dex-induced B-ALL death. Good agreement was observed among 3 biological replicates, and the shRNAs show remarkably consistent activities within any given gene (supplemental Figure 4A-D). Importantly, genes that, when knocked down, either significantly protect (resulting in shRNA enrichment) or sensitize (resulting in shRNA depletion) NALM-6 cells to dex are evident (Figure 2B; supplemental Document 3). Overall, knockdown of 156 of 5761 CAGEK genes had highly significant effects (false discovery rate, ≤5%) on NALM-6 dex sensitivity, and those we tested individually validated well (supplemental Figure 4E-F). About 10% of the genes screened (653) had an effect on NALM-6 growth. In addition to these highly significant genes, a number of other genes (P ≤ .05), which we term top hits, likely also affect GC sensitivity (Figure 2B; supplemental Figure 5B).
It should be noted that although knockdown of GR resulted in complete resistance, protective hits from the screen generally decreased sensitivity only twofold to threefold (supplemental Figure 4E). This could be the result of incomplete knockdown (supplemental Figure 4F) or compensation by other factors. However, an alternative model is that GC-induced cell death is multifactorial, having multiple downstream effectors of cytotoxicity, and a network of signals upstream that collaborate with GR to efficiently induce cell death. This model is supported by the identification of many unexpected modulators of dex sensitivity (Figure 2B; supplemental Figure 6A-B). For example, the screen not only confirmed the importance of BCL2, which was sensitizing upon knockdown, and BCL2L11 and TXNIP, which were protective, but also identified 4 other key BH3-containing factors that affected dex-induced cell death (supplemental Figure 6C). This demonstrates that no apoptosis gene is absolutely required. Furthermore, the partial effects of these genes reinforce the idea that multiple factors contribute to GC cytotoxicity.
The screen also revealed important new insights into the cellular factors that affect GC cytotoxicity and sensitivity. Among the genes screened are 21 genes that are frequently mutated in refractory/relapsed B-ALL, 16 (∼70%) of which are among our top hits (Figure 2C; supplemental Table 1).1,2 Most of these genes are sensitizing when knocked down, suggesting that rare gain-of-function mutations conferring resistance to GCs are selected for during treatment. These data demonstrate that perturbing GC function is perhaps the most prevalent source of overall treatment resistance.
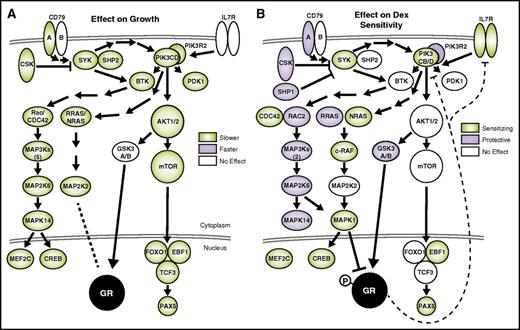
Most strikingly, pathway analysis of hits affecting GC sensitivity revealed a role for B-cell development in GC cytotoxicity. The BCR pathway (supplemental Figure 3D) has the most significant effect on both the growth and sensitivity of NALM-6 cells to dex. The BCR pathway is a potent growth and survival signal that works in part through stimulation of the phosphatidylinositol 3-kinase (PI3K) and extracellular signal-regulated kinase (ERK)/MAPK pathways,25 knockdown of which also exhibited significant effects on growth and sensitivity (mapped in Figure 3). The mechanism of how BCR/PI3K signaling affects GC cytotoxicity is revealed by integrating the functional genomic and gene expression data.
Suppression of BCR signaling is detrimental to growth and sensitizes B-ALL to dex. The effects of gene knockdown on growth (A) and dex sensitivity (B) are overlaid on components of the BCR pathway. Genes are present when included in the screen, and shaded when the effect of knockdown is significant (Mann-Whitney, P ≤ .05). Dashed lines indicate repression of PIK3CD and IL7R expression by dex (diagrams based on Ingenuity pathways, and other literature25,26 ).
Suppression of BCR signaling is detrimental to growth and sensitizes B-ALL to dex. The effects of gene knockdown on growth (A) and dex sensitivity (B) are overlaid on components of the BCR pathway. Genes are present when included in the screen, and shaded when the effect of knockdown is significant (Mann-Whitney, P ≤ .05). Dashed lines indicate repression of PIK3CD and IL7R expression by dex (diagrams based on Ingenuity pathways, and other literature25,26 ).
One challenge in interpreting differential gene expression data sets is identifying which regulated genes cause the phenotype, in this case cell death. We used the screen to identify these “effector genes” from among the CRGs en masse. Dex-activated effector genes (those that induce cell death when activated) increase growth or protect cells from dex-induced cell death when knocked down. Conversely, knockdown of dex-repressed effector genes (those whose repression induces cell death) decreases growth or increases sensitivity to dex. Of the CRGs included in the screen (247 of 588), 85 of those knocked down caused a growth phenotype, including 56 that match the effector phenotype (supplemental Figures 5A,C and 7A). Several of the activated effector genes are transcriptional cofactors, including BTG1, which is required for GR autoinduction, a consistent feature of dex-sensitive B-ALL.27 A larger number of repressed genes exhibited an effector phenotype, including key regulators of lymphoid and B-cell development (MEF2C/D, LEF1, RUNX1, ETV6, BCL2, and TCF4; supplemental Figure 7A), supporting our model that GC regulation of B-cell development genes contributes to its cytotoxicity.
The importance of B-cell development pathways in GC cytotoxicity is even more evident from analysis of effector genes identified by dex sensitivity in the screen. Of the 63 GC-regulated genes whose knockdown affects dex sensitivity, about half (32) exhibited an effector phenotype (Figure 2A,D; supplemental Figure 7B). Genes that are activated and protective are overrepresented for transcription-related factors (GR, SERTAD1, SMARCA2, CTBP1, SBF1, STK40), which, like BTG1, may be required for GR gene regulation or to enhance downstream transcriptional programs. A striking number of repressed and sensitizing effector genes are involved in lymphoid and B-cell development, including BCL2, LEF1, IL7R, CBX4, CMTM7, ZMIZ1, TCF4, and PIK3CD. This not only supports the link between development and GC efficacy, but also suggests that these synthetic interactions can be exploited with inhibitors to synergize with GCs.
PI3Kδ and the BCR pathway are tightly regulated by GCs
An intimate connection between GCs and B-cell development is evident in the BCR pathway. Tonic signaling through the pre-BCR is present in ∼15% of B-ALL, including NALM-6 cells, and is essential for B-cell development and survival.28 Indeed, our screen data indicate that knockdown of almost any BCR component is detrimental to growth, confirming the importance of the pathway (Figure 3A). In contrast, only the PI3K/RAS/MAPK branch of the BCR pathway sensitizes cells to dex (Figure 3B). Thus, pre-BCR signaling not only drives proliferation of B-ALL cells, but it also specifically opposes dex-induced cell death.
The importance of BCR signaling in treatment sensitivity of B-ALL has been shown previously through inhibition of the mammalian target of rapamycin (mTOR)/AKT branch with rapamycin.29 Although we also observe an effect of mTOR/AKT knockdown on growth, it does not sensitize B-ALL to GCs. This is in contrast to T-cell ALL, where AKT inhibition does synergize with dex.30 Instead, our data highlight the PI3K/MAPK branch, which can be activated from the BCR proximal SYK or from IL7R, which converge specifically on the lymphoid-restricted PI3Kδ (PIK3CD), through NRAS, eventually inhibiting GR function through phosphorylation by Erk2 (MAPK1) (Figure 3B).
Tight control of specific PI3K signaling components by GCs is apparent from the gene expression and shRNA screen data (Figure 4A). Of the p110 PI3K subunits, knockdown of only PIK3CD both inhibited cell growth and sensitized cells to dex (Figure 4B), whereas knockdown of PTEN protected cells from dex (Figure 4C). A specific regulatory mechanism is evident, as knockdown of only one p85 regulatory subunit, PIK3R2, protected cells from dex-induced cell death (Figure 4C), consistent with its role in restraining production of phosphatidylinositol 3,4,5 trisphosphate.31 Not only do these specific PI3K components affect GC sensitivity, but their expression is also regulated by dex. Expression of PI3Kδ is strongly repressed by dex, and PIK3IP1, a negative regulator of PI3Ks,32 is strongly activated (Figure 4E left). The presence of GR-binding sites in sensitive cells for both of these genes indicates direct regulation by dex (Figure 4E right). Together, these data map a double-negative feedback loop between PI3Kδ and GR: addition of GCs suppresses PI3K activity, which in turn sensitizes cells to GCs (Figure 4A).
Disruption of double-negative feedback loop between PI3Kδ and GR enhances dex cytotoxicity. (A) Schematic feedback loop based on combined data from the shRNA screen and microarray gene expression data. Dex-induced repression of PIK3CD (blue blocking arrow, PI3Kδ) and activation of PIK3IP1 (red arrow) gene expression. shRNA knockdown of PTEN and PIK3R2 was protective (purple), whereas knockdown specifically of PIK3CD sensitized cells to dex (green). Thus, interruption of PIK3δ inhibition of GR is expected to synergistically induce cell death. (B-D) Results of the shRNA screen. Bar graphs show the log10(P) of the hits from the shRNA screen. Sensitizing hits have been depicted as negative (green), protective as positive (purple). (E) Effect of dex on gene expression. Fold change of gene expression across sensitive B-ALL samples as measured by microarray (left) and GR occupancy as measured by ChIP-seq (right) post-dex treatment. Primary and PDX samples are marked red; cell lines, black. ChIP-seq data are shown for sensitive (B1) and resistant (HM3101) samples. The presence of GR-binding sites in sensitive cells for both PIK3IP1 and PIK3CD indicates potential direct regulation by dex. (F) The combination index of dex and CAL-101 in sensitive (NALM-6, SUP-B15) and resistant (RCH-ACV) cell lines, a resistant patient sample (HM3101), and a multiply relapsed refractory patient-derived mouse xenograft (ALL121) (super additive <1; CalcuSyn). Numbers reflect isobolograms depicted in supplemental Figure 9. (G) Quantification of westerns against phospho-S203 of GR in the absence and presence of PI3Kδ inhibition (error bars represent standard error of the mean [SEM] across 4 time points). CAL-101 treatment reduces GR S203 phosphorylation, likely increasing GR activity. (H) Spleens of mice (n = 5 mice/cohort) engrafted with relapsed B-ALL cells (ALL121) and treated with vehicle, dex (7.5 mg/kg), idela (50 mg/kg), or both for 2 weeks. Enlarged spleens indicate the accumulation of lymphoblasts. Treatment with either dex or idela alone failed to significantly reduce spleen size compared with untreated control; however, treatment with both dex and idela significantly reduced spleen size, indicating a synergistic effect between the 2 drugs. (I) Total number of human ALL cells (y-axis) in spleens of mice in panel H as measured by quantitative flow cytometry.
Disruption of double-negative feedback loop between PI3Kδ and GR enhances dex cytotoxicity. (A) Schematic feedback loop based on combined data from the shRNA screen and microarray gene expression data. Dex-induced repression of PIK3CD (blue blocking arrow, PI3Kδ) and activation of PIK3IP1 (red arrow) gene expression. shRNA knockdown of PTEN and PIK3R2 was protective (purple), whereas knockdown specifically of PIK3CD sensitized cells to dex (green). Thus, interruption of PIK3δ inhibition of GR is expected to synergistically induce cell death. (B-D) Results of the shRNA screen. Bar graphs show the log10(P) of the hits from the shRNA screen. Sensitizing hits have been depicted as negative (green), protective as positive (purple). (E) Effect of dex on gene expression. Fold change of gene expression across sensitive B-ALL samples as measured by microarray (left) and GR occupancy as measured by ChIP-seq (right) post-dex treatment. Primary and PDX samples are marked red; cell lines, black. ChIP-seq data are shown for sensitive (B1) and resistant (HM3101) samples. The presence of GR-binding sites in sensitive cells for both PIK3IP1 and PIK3CD indicates potential direct regulation by dex. (F) The combination index of dex and CAL-101 in sensitive (NALM-6, SUP-B15) and resistant (RCH-ACV) cell lines, a resistant patient sample (HM3101), and a multiply relapsed refractory patient-derived mouse xenograft (ALL121) (super additive <1; CalcuSyn). Numbers reflect isobolograms depicted in supplemental Figure 9. (G) Quantification of westerns against phospho-S203 of GR in the absence and presence of PI3Kδ inhibition (error bars represent standard error of the mean [SEM] across 4 time points). CAL-101 treatment reduces GR S203 phosphorylation, likely increasing GR activity. (H) Spleens of mice (n = 5 mice/cohort) engrafted with relapsed B-ALL cells (ALL121) and treated with vehicle, dex (7.5 mg/kg), idela (50 mg/kg), or both for 2 weeks. Enlarged spleens indicate the accumulation of lymphoblasts. Treatment with either dex or idela alone failed to significantly reduce spleen size compared with untreated control; however, treatment with both dex and idela significantly reduced spleen size, indicating a synergistic effect between the 2 drugs. (I) Total number of human ALL cells (y-axis) in spleens of mice in panel H as measured by quantitative flow cytometry.
To test this hypothesis, we inhibited PI3Kδ using CAL-101 (idelalisib [idela]), a US Food and Drug Administration–approved drug used in monotherapy treatment of chronic lymphocytic leukemia and indolent non-Hodgkin lymphoma.33 Although CAL-101/idela monotherapy shows an effect in PDX models of treatment-refractory pediatric B-ALL, it is does not clear the disease.34 We tested the combination of CAL-101 and dex in 5 B-ALL samples: 2 sensitive cell lines (NALM-6, SUP-B15) and 3 resistant samples (cell line RCH-ACV, relapsed patient sample HM3101, and relapsed PDX, ALL121). The response of cells to CAL-101 was different than to dex, with SUP-B15 being the least sensitive and RCH-ACV being the most sensitive (supplemental Figure 8A-B). Graphing the isoboles35 revealed that dex and CAL-101 are superadditive in all samples, including the most refractory B-ALL patient samples (supplemental Figure 9). With a combination index as low as 0.13 (Figure 4F), the superadditivity in all backgrounds indicates that addition of CAL-101 sensitizes B-ALL to dex and may be effective in overcoming resistance. This synergy, surprisingly, is independent of pre-BCR status36 suggesting that PI3Kδ may be activated by IL7R or other pathways in the absence of tonic pre-BCR signaling. Thus, although synergy had also been observed with a pan-PI3K inhibitor,37 the resolution of our data allow targeting of the lymphoid-restricted PI3Kδ, which is likely to have fewer side effects.
As proof of concept, we tested this combination in a PDX model of ALL. NOD scid γ-deficient (NSG) mice engrafted with leukemia cells from a child with multiply relapsed B-ALL (ALL121, Ph-like, pre-BCR−) were treated with vehicle, dex (7.5 mg/kg daily), idela (50 mg/kg daily), or both for 2 weeks. Harvested spleens from treated animals demonstrated no effect from dex or idela monotherapy, but markedly reduced spleen size and decreased human leukemia burden using the dex/idela combination (Figure 4H-I). The combination shows a more pronounced synergy in this model than predicted from in vitro cultures. Although more preclinical work is needed to determine how prevalent the efficacy of this combination is, the PDX model shows the utility of this integrated functional genomic approach in identifying promising combination chemotherapeutics.
PI3Kδ inhibition potentiates regulation of effector genes
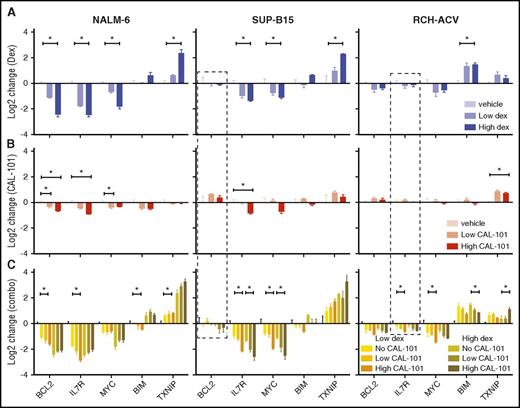
The synergy of dex and CAL-101/idela is due, at least in part, to enhanced GC-regulation of effector genes. Using combinations of dex and CAL-101, we monitored 4 repressed (BCL2, IL7R, MYC, PIK3CD) and 2 activated (BCL2L11 and TXNIP) effector genes in 3 cell lines: NALM-6 (sensitive to both drugs), SUP-B15 (sensitive to dex but resistant to CAL-101), and RCH-ACV (resistant to dex and sensitive to CAL-101) (Figure 5). Inhibition of PI3Kδ significantly enhanced dex-induced repression of BCL2 and IL7R (at low doses of dex), and activation of TXNIP in all cells tested (Figure 5). Surprisingly, inhibition of PI3Kδ had little, or even an opposing, effect on dex repression of PIK3CD (supplemental Figure 10D), indicating that inhibition of PI3Kδ does not feed back on itself through GR. For other effector genes, the combined regulation is cell-type specific, exemplified by the effect of PI3Kδ inhibition on MYC repression, which is enhanced in SUP-B15 and RCH-ACV cells, but not in NALM-6. In addition, dex-induced activation of BIM, thought to be a crucial component of dex-induced B-ALL cell death, is blunted by PI3Kδ inhibition, again suggesting that other BH3 family members may be important in driving apoptosis. This potentiation can work directly through GR at genes such as TXNIP in NALM-6 cells, where CAL-101 alone has no effect on regulation yet enhances dex-induced activation. Potentiation can also be combinatorial for some genes, as is the case with MYC in RCH-ACV cells: CAL-101 and dex both regulate the gene in the same direction, but they regulate more strongly together. These data indicate that inhibition of PI3Kδ synergizes with dex in a cell-type–specific manner by selectively potentiating regulation of different sets of effector genes.
Inhibition of PI3Kδ synergizes with dex in regulating cell-death effector genes. (A) Change in gene expression measured by quantitative polymerase chain reaction (qPCR) in response to 2 concentrations of dex at 24 hours in 3 cell lines. (B) Change in gene expression measured by qPCR in response to 2 concentrations of CAL-101 alone and in combination with 2 concentrations of dex (C) at 24 hours in the same cell lines. Experiments represent at least 3 biological repeats. *P ≤ .05 (see “Methods” for details). Dashed boxes highlight genes whose regulation is restored by CAL-101 (idela).
Inhibition of PI3Kδ synergizes with dex in regulating cell-death effector genes. (A) Change in gene expression measured by quantitative polymerase chain reaction (qPCR) in response to 2 concentrations of dex at 24 hours in 3 cell lines. (B) Change in gene expression measured by qPCR in response to 2 concentrations of CAL-101 alone and in combination with 2 concentrations of dex (C) at 24 hours in the same cell lines. Experiments represent at least 3 biological repeats. *P ≤ .05 (see “Methods” for details). Dashed boxes highlight genes whose regulation is restored by CAL-101 (idela).
CAL-101/idela administration can also restore GC-induced regulation of quiescent genes. BCL2 in SUP-B15 cells and IL7R in RCH-ACV cells do not respond to dex alone, but when treated with a combination of CAL-101 and dex, they are repressed (Figure 5 dashed boxes). This result supports the model that resistance to GCs can be due to a failure of GR to regulate key genes, and, further, that a latent GC-regulated cell death program can be re-established by manipulation of key pathways that converge on GR. To test convergence, we inhibited PI3Kδ and probed 3 phosphorylation sites on GR known to modulate its function: S203, S211, and S226. Administration of CAL-101 specifically reduces S203 phosphorylation levels (Figure 4G), which has been shown to inhibit GR function.38,39 CDK2A can phosphorylate S203,40,41 but our screen indicates that only MAPK1 has an effect on GC sensitivity, indicating that the PI3K/MAPK pathway inhibits specific GR functions.
Discussion
The link between B-cell development and cytotoxicity suggests that GCs perform a function in normal B-cell development as 1 of many signals that influence B-cell selection. This connection echoes the positive and negative effect of GCs in early T-cell development,42 and is reminiscent of the therapeutic effect of all-trans retinoic acid on another nuclear hormone receptor, the retinoic acid receptor, whose cytotoxic effect on acute promyelocytic leukemia results from pushing through a developmental block.43 The ability of GCs to suppress B-cell checkpoint genes that are required across multiple developmental stages helps explain their efficacy in treating lymphoid malignancies that are blocked at these different stages. Indeed, in NALM-6 cells, knockdown of the BCR pathway and IL7R enhances dex-induced death, suggesting arrest at the pre-B stage, which is consistent with the genetic and cytological features of these cells.44 We therefore propose the model that GCs can either stop or push B cells through development (Figure 1C). In this model, supraphysiological levels of GCs can either push immature cells to the next stage of development (through BCL6 or CXCR4, for example), which may trigger apoptotic programs, or they may arrest cells by removing a positive growth signal (such as IL7R, PIK3CD, or ITGA4). In the case of IL7R and PIK3CD, gene repression may further accentuate GR function, forming a positive feedback loop that drives cells toward death. Further study is needed to validate this model, but it provides a long-sought mechanism for how and why GCs induce lymphoid cell death.
The resolution of next-generation shRNA screening establishes it as an essential tool for rational identification of combination chemotherapeutics.45 Hits can be filtered for potential synergy, tissue restriction, and for the availability of existing drugs. Using these criteria, we demonstrate a pipeline to rapidly identify potent combination therapies that are likely to have fewer side effects and accelerated time to preclinical and clinical testing.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by National Institutes of Health, National Cancer Institute awards K99/R00 CA149088 (M.A.P.), K99/R00 CA181494 (M.K.), and K08CA184418 (S.K.T.), a Stand Up to Cancer Innovative Research grant (M.K.), the V Foundation (M.A.P.), and the Roy J. Carver Charitable Trust 01-224 (M.A.P.).
Authorship
Contribution: M.A.P. conceived of, wrote, and performed the bioinformatics analysis; K.A.K., M.F., D.N.S., and O.A.-H. performed the experimental work; R.M. identified PI3Kδ and performed initial experiments thereon; H.Y. performed some key bioinformatic analyses; S.K.T. provided samples, insight, and performed the PDX mouse experiments; M.L.L. provided insight and samples; M.M. assisted conceptually and with writing; M.C.B. and J.S.W. were essential in setting up the shRNA screen; and M.K. was a close collaborator in the design, execution, processing, and interpretation of the shRNA screen and preparing the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Miles A. Pufall, Department of Biochemistry, University of Iowa, 51 Newton Rd, BSB 4-430, Iowa City, IA 52242; e-mail: miles-pufall@uiowa.edu.


![Figure 4. Disruption of double-negative feedback loop between PI3Kδ and GR enhances dex cytotoxicity. (A) Schematic feedback loop based on combined data from the shRNA screen and microarray gene expression data. Dex-induced repression of PIK3CD (blue blocking arrow, PI3Kδ) and activation of PIK3IP1 (red arrow) gene expression. shRNA knockdown of PTEN and PIK3R2 was protective (purple), whereas knockdown specifically of PIK3CD sensitized cells to dex (green). Thus, interruption of PIK3δ inhibition of GR is expected to synergistically induce cell death. (B-D) Results of the shRNA screen. Bar graphs show the log10(P) of the hits from the shRNA screen. Sensitizing hits have been depicted as negative (green), protective as positive (purple). (E) Effect of dex on gene expression. Fold change of gene expression across sensitive B-ALL samples as measured by microarray (left) and GR occupancy as measured by ChIP-seq (right) post-dex treatment. Primary and PDX samples are marked red; cell lines, black. ChIP-seq data are shown for sensitive (B1) and resistant (HM3101) samples. The presence of GR-binding sites in sensitive cells for both PIK3IP1 and PIK3CD indicates potential direct regulation by dex. (F) The combination index of dex and CAL-101 in sensitive (NALM-6, SUP-B15) and resistant (RCH-ACV) cell lines, a resistant patient sample (HM3101), and a multiply relapsed refractory patient-derived mouse xenograft (ALL121) (super additive <1; CalcuSyn). Numbers reflect isobolograms depicted in supplemental Figure 9. (G) Quantification of westerns against phospho-S203 of GR in the absence and presence of PI3Kδ inhibition (error bars represent standard error of the mean [SEM] across 4 time points). CAL-101 treatment reduces GR S203 phosphorylation, likely increasing GR activity. (H) Spleens of mice (n = 5 mice/cohort) engrafted with relapsed B-ALL cells (ALL121) and treated with vehicle, dex (7.5 mg/kg), idela (50 mg/kg), or both for 2 weeks. Enlarged spleens indicate the accumulation of lymphoblasts. Treatment with either dex or idela alone failed to significantly reduce spleen size compared with untreated control; however, treatment with both dex and idela significantly reduced spleen size, indicating a synergistic effect between the 2 drugs. (I) Total number of human ALL cells (y-axis) in spleens of mice in panel H as measured by quantitative flow cytometry.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/129/22/10.1182_blood-2017-02-766204/4/m_blood766204f4.jpeg?Expires=1765901981&Signature=zRTELSxonfuqaTaElJfs-uI3d2J4NoZ1lAsL5-aQ~lhpH1K3zQfo1V9~SOWlSUBh5SO6OSfZ7fnmoYzj-IH6-bZWLoyg6NAp2t8Eq4~7Kl~MWMndeb0yTvah7ULwj1OWs2-889Y-O54WWNQTWGvi67NseaAwaqOnLFO6OVsvRIfXMd-H-RBdlnoecvFR4xMSrja-N~z6VtpRfoORgd5AhuR3ULJdpcL30pNxa7lSpogGb68Jl0VGca6G2K51xbcyFTxsmsZR9zy62JEzBcbKtcCFaQwmJN43et249f6wLoVtpOKUDWHE38Q9Xcm-u8JnP1Yiz9mv5yqUXFI4BJijSA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal