Key Points
We validated the association of a circulating genome-wide gene expression profile with poor outcomes in 3 cohorts of SCD.
A composite risk score using this genomic biomarker with clinical risk factors exhibited improved prediction than clinical factors alone.
Abstract
Sickle cell disease (SCD) complications are associated with increased morbidity and risk of mortality. We sought to identify a circulating transcriptomic profile predictive of these poor outcomes in SCD. Training and testing cohorts consisting of adult patients with SCD were recruited and prospectively followed. A pathway-based signature derived from grouping peripheral blood mononuclear cell transcriptomes distinguished 2 patient clusters with differences in survival in the training cohort. These findings were validated in a testing cohort in which the association between cluster 1 molecular profiling and mortality remained significant in a fully adjusted model. In a third cohort of West African children with SCD, cluster 1 differentiated SCD severity using a published scoring index. Finally, a risk score composed of assigning weights to cluster 1 profiling, along with established clinical risk factors using tricuspid regurgitation velocity, white blood cell count, history of acute chest syndrome, and hemoglobin levels, demonstrated a higher hazard ratio for mortality in both the training and testing cohorts compared with clinical risk factors or cluster 1 data alone. Circulating transcriptomic profiles are a powerful method to risk-stratify severity of disease and poor outcomes in both children and adults, respectively, with SCD and highlight potential associated molecular pathways.
Introduction
Sickle cell disease (SCD) is characterized by chronic hemolytic anemia, vaso-occlusion, and multiorgan dysfunction. Contributing pathways to the development of these complications including the role of hemolysis and heme-based injury, the vascular endothelium, the effects of hypoxia and inflammation- and immune-mediated cell activation have been extensively studied.1-7 Despite its Mendelian inheritance, the course of patients with SCD is highly variable. There is significant heterogeneity in the rate of development of acute and chronic pain, cerebrovascular disease, acute chest syndrome,8,9 pulmonary hypertension (PH), diastolic dysfunction, renal failure, hemolytic anemia, and premature or sudden death.10-13 Therefore, the development of biomarkers for risk stratification is vital. The utility of such biomarkers in the management of SCD is underscored by their potential application as tools to provide early diagnosis of complications, to identify a subset of individuals at risk for a severe clinical course, to define disease relevant molecular pathways, or to monitor response to therapy.
More than 100 different biomarkers have been described in SCD,14 and nearly all target a particular molecule, gene, or manifestation of SCD associated with poor outcomes, with a small subset that have been evaluated in both children and adults. Evidence suggests that plasma concentrations of targeted proteins or changes in blood cells may be informative of disease presence, severity, and prognosis.14 Changes in the peripheral blood transcriptome of patients with SCD have been well documented7,15-20 ; however, the ability of the transcriptome to predict outcomes has not been assessed. Given the established role of peripheral blood mononuclear cells (PBMCs) in inflammation and the utility of PBMC-derived gene expression in both diagnostic and prognostic aspects of other entities,21-25 we hypothesized that PBMC gene expression profiling may be predictive of poor outcomes in patients with SCD. We sought to define a circulating molecular biomarker that would further risk-stratify all patients with SCD and represent a convergence of several etiologies leading to poor outcomes.
Methods
Study design and cohorts
All subjects in all cohorts were prospectively recruited and provided written consent to participate in this study with the approval by the respective institutional human subjects review boards. The training cohort consisted of patients prospectively recruited from the University of Illinois (n = 172) since 2010. The testing cohort (n = 78) consisted of patients prospectively seen at the University of Chicago (n = 38) and Howard University (n = 40) since 2007. All patients were consecutively seen and recruited from outpatient clinics in steady-state conditions (no vaso-occlusive crises in 3 weeks; further defined in the supplemental Methods, available on the Blood Web site). Details of the West African children cohort were previously reported, including a description of clinical severity26 using a severity score derived from an online calculator (http://www.bu.edu/sicklecell/projects/).27 Each patient in this latter cohort was assigned a score based on sex, hemoglobin (Hb) genotype, mean corpuscular volume, and white blood cell (WBC) counts; whole blood was used for gene expression profiling.
Pathway signature and analysis
Details of microarray preparation and methods including consensus clustering are provided in the supplemental methods. The Functional Analysis of Individual Microarray Expression (FAIME) is an algorithm to compute pathway scores using rank-weighted gene expression of an individual sample.28,29 FAIME has been demonstrated to produce more reliable validation than gene signature predictors in several studies.30,31 Based on the FAIME signatures (supplemental methods), support vector machine (SVM) classifiers were developed and used to classify the observed expression clusters.
Gene signature
Based on the clusters identified in the training cohort, we derived separate gene signatures (fold change, ≥1.5; false discovery rate [FDR], ≤1 × 10−5) differentiating clusters 1 and 2 in the training and testing cohorts. Overlap between these 2 gene signatures identified a 31-gene signature associated with cluster-specific profiles in SCD. An SVM classifier based on this derived gene signature was constructed and tested in the West African cohort.
Risk score
A risk score was constructed integrating clinical markers of risk and transcriptomic data (cluster profiling). Clinical markers of risk were chosen based on established markers of severity of and mortality in SCD including an elevated tricuspid regurgitation velocity (TRV, ≥2.5 m/s),30 history of acute chest syndrome (ACS),8 elevated WBC count (≥103/μL),31 and reduced Hg levels (≤10 g/dL).32 Each clinical variable and cluster identification (with cluster 1 representing high risk) was assigned a value of 0 for low risk and 1 for high risk based on whether the values crossed the given thresholds. A composite risk score of ≥4 was considered high risk when integrating all individual variables because it was associated with the greatest mortality risk. The patients were stratified into 2 groups (ie, high risk and low risk) based on the ability of the composite risk score to demonstrate the greatest survival difference.
Statistics
Results are presented as mean and standard deviation with Student t test of participants with a given characteristic. Fisher exact test was used for categorical values such as sex and use of hydroxyurea. Proportional hazards (Cox) regression was used to study relationships between covariates of interest and mortality. The time-to-event outcome analyzed was vital status from blood draw until death or completion of the study on April 1, 2015, and determined by a combination of social security death index, follow-up calls, and review of electronic medical record. The risk ratio (hazard ratio [HR]) and 95% confidence interval (CI) for each predictor were determined and Kaplan-Meier survival curves were calculated. Because this was a registry study, prospective power analysis was not performed. All analyses were performed using R (version 3.0.1) software (https://www.r-project.org/). P or FDR <.05 was considered statistically significant.
Results
Unsupervised consensus clustering
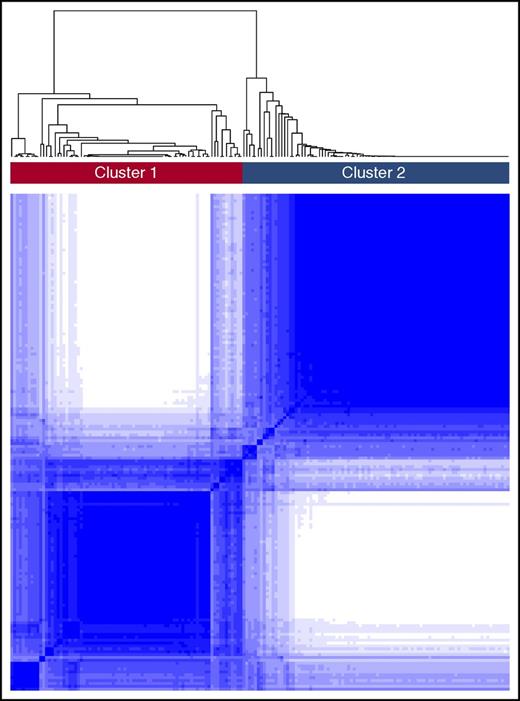
A consensus matrix heat map from unbiased consensus clustering and a plot of the cumulative distribution function (CDF) of consensus indices revealed the presence of 2 distinct global transcriptomic subtypes or clusters from subjects in the training cohort (labeled clusters 1 and 2, Figure 1). To determine this optimal number of clusters, the change in area under the CDF was evaluated in response to increasing the number of clusters, K. When K increased from 2 to 3, the area under the CDF did not markedly increase. In addition, the consensus matrix heat map for K = 3 demonstrated a high proportion of samples with ambiguous clustering (supplemental Figure 1). These provided evidence of the optimal 2 transcriptomic clusters.
Consensus clustering in SCD. The figure depicts the matrix of all consensus indices with each element in this matrix representing the index for 1 pair of samples. In an ideal matrix, all consensus indices would be 1 or 0, indicating that each pair of samples always or never, respectively, clustered together during the resampling. In the training cohort, 2 broad clusters were identified upon resampling.
Consensus clustering in SCD. The figure depicts the matrix of all consensus indices with each element in this matrix representing the index for 1 pair of samples. In an ideal matrix, all consensus indices would be 1 or 0, indicating that each pair of samples always or never, respectively, clustered together during the resampling. In the training cohort, 2 broad clusters were identified upon resampling.
Table 1 provides demographic and clinical information from the cohorts, differentiated by clusters 1 and 2. Most patients in the training and testing cohorts had HgSS genotype. Although no significant differences were present across several variables, in the training cohort, patients in cluster 1 had significantly elevated WBC counts, total bilirubin levels, increased prevalence of reported history of ACS, and lower Hg levels compared with patients in cluster 2. In the testing cohort, WBC levels were also higher in cluster 1 patients compared with those of cluster 2, whereas the remaining variables (elevated bilirubin, LDH) trended in a similar direction. Interestingly, elevated aspartate aminotransferase (AST) levels in cluster 1 were observed in both cohorts, reaching near significance. In both cohorts, basic demographics such as age, sex, and BMI and use of hydroxyurea as well as peak TRV (measurements based on echocardiography following American Society of Echocardiography guidelines) were not significantly different between clusters 1 and 2.
Clinical and demographic characteristics of the training and testing cohorts stratified by cluster
| . | Training cohort . | Testing cohort . | ||||
|---|---|---|---|---|---|---|
| . | Cluster 1 (n = 80) . | Cluster 2 (n = 92) . | P value . | Cluster 1 (n = 35) . | Cluster 2 (n = 37) . | P value . |
| Age (y) | 34.8 ± 11.8 | 36.3 ± 12.0 | .41 | 38.5 ± 9.1 | 38.2 ± 11.8 | .91 |
| Female/male (n) | 43/37 | 51/41 | .09 | 17/22 | 28/22 | .29 |
| HgSS | 71 | 76 | .35 | 29 | 31 | .76 |
| HgSC | 5 | 12 | .20 | 4 | 7 | .749 |
| HgSB | 4 | 4 | 1.00 | 2 | 3 | 1.00 |
| BMI (kg/m2) | 25.1 ± 6.0 | 24.8 ± 5.3 | .74 | 22.8 ± 3.8 | 24.2 ± 6.6 | .28 |
| HU therapy | 40 (50%) | 48 (52%) | .88 | 16 (53%) | 27 (69%) | .21 |
| ACS | 73 (91%) | 71 (77%) | .013 | 23 (62%) | 20 (45%) | .18 |
| WBC (10−3/mm3) | 10.7 ± 3.6 | 9.0 ± 3.4 | .002 | 10.2 ± 3.5 | 8.6 ± 3.3 | .039 |
| Hg (g/dL) | 8.5 ± 1.6 | 9.1 ± 1.8 | .045 | 8.7 ± 2.0 | 8.8 ± 1.7 | .77 |
| Creatinine (mg/dL) | 0.98 ± 0.83 | 1.22 ± 1.58 | .22 | 1.02 ± 1.38 | 1.27 ± 1.80 | .47 |
| HbF (g/dL) | 7.1 ± 5.3 | 8.0 ± 7.6 | .39 | 8.5 ± 8.7 | 9.4 ± 7.8 | .73 |
| LDH (U/L) | 453 ± 222 | 394 ± 195 | .07 | 378 ± 170 | 350 ± 154 | .50 |
| Bilirubin (mg/dL) | 3.9 ± 3.1 | 2.3 ± 1.5 | .0001 | 2.8 ± 1.6 | 2.2 ± 1.6 | .13 |
| AST (U/L) | 57.0 ± 69.3 | 41.5 ± 21.2 | .060 | 47.1 ± 27.4 | 37.4 ± 17.0 | .07 |
| Peak TRV (≥2.5 m/s) | 39.1% | 26.3% | .109 | 60.0% | 50.0% | .487 |
| Peak TRV (≥3.0 m/s) | 12.5% | 5.0% | .134 | 14.3% | 7.5% | .461 |
| . | Training cohort . | Testing cohort . | ||||
|---|---|---|---|---|---|---|
| . | Cluster 1 (n = 80) . | Cluster 2 (n = 92) . | P value . | Cluster 1 (n = 35) . | Cluster 2 (n = 37) . | P value . |
| Age (y) | 34.8 ± 11.8 | 36.3 ± 12.0 | .41 | 38.5 ± 9.1 | 38.2 ± 11.8 | .91 |
| Female/male (n) | 43/37 | 51/41 | .09 | 17/22 | 28/22 | .29 |
| HgSS | 71 | 76 | .35 | 29 | 31 | .76 |
| HgSC | 5 | 12 | .20 | 4 | 7 | .749 |
| HgSB | 4 | 4 | 1.00 | 2 | 3 | 1.00 |
| BMI (kg/m2) | 25.1 ± 6.0 | 24.8 ± 5.3 | .74 | 22.8 ± 3.8 | 24.2 ± 6.6 | .28 |
| HU therapy | 40 (50%) | 48 (52%) | .88 | 16 (53%) | 27 (69%) | .21 |
| ACS | 73 (91%) | 71 (77%) | .013 | 23 (62%) | 20 (45%) | .18 |
| WBC (10−3/mm3) | 10.7 ± 3.6 | 9.0 ± 3.4 | .002 | 10.2 ± 3.5 | 8.6 ± 3.3 | .039 |
| Hg (g/dL) | 8.5 ± 1.6 | 9.1 ± 1.8 | .045 | 8.7 ± 2.0 | 8.8 ± 1.7 | .77 |
| Creatinine (mg/dL) | 0.98 ± 0.83 | 1.22 ± 1.58 | .22 | 1.02 ± 1.38 | 1.27 ± 1.80 | .47 |
| HbF (g/dL) | 7.1 ± 5.3 | 8.0 ± 7.6 | .39 | 8.5 ± 8.7 | 9.4 ± 7.8 | .73 |
| LDH (U/L) | 453 ± 222 | 394 ± 195 | .07 | 378 ± 170 | 350 ± 154 | .50 |
| Bilirubin (mg/dL) | 3.9 ± 3.1 | 2.3 ± 1.5 | .0001 | 2.8 ± 1.6 | 2.2 ± 1.6 | .13 |
| AST (U/L) | 57.0 ± 69.3 | 41.5 ± 21.2 | .060 | 47.1 ± 27.4 | 37.4 ± 17.0 | .07 |
| Peak TRV (≥2.5 m/s) | 39.1% | 26.3% | .109 | 60.0% | 50.0% | .487 |
| Peak TRV (≥3.0 m/s) | 12.5% | 5.0% | .134 | 14.3% | 7.5% | .461 |
BMI, body mass index; HbF, hemoglobin F; HU, hydroxyurea; LDH, lactate dehydrogenase.
When stratified by cohorts rather than clusters (supplemental Table 1), the testing cohort had a slightly lower BMI (clinically insignificant), AST, and LDH values than the training cohort. However, the testing cohort subjects were older in age, with higher averages of peak TRV ≥2.5 m/s values but not ≥3.0 m/s values. The lack of difference in the latter values, which represent a greater likelihood of having right heart catheterization–confirmed PH, support the notion that the presence of PH between the testing and training cohorts was likely not significantly different.
Consensus clustering and mortality
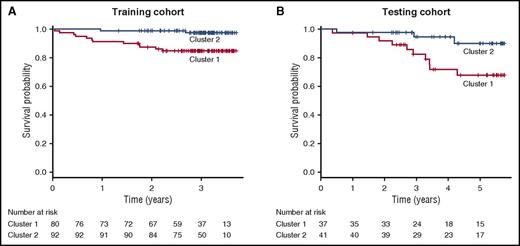
During a median follow-up of 3.07 years (95% CI, 2.98-3.15), 8.1% of patients (n = 14) died in the training cohort. Twelve of the patients that died and 68 of those that survived exhibited cluster 1 molecular profiling, whereas cluster 2 molecular profiling was present in 2 of the patients that died and 90 patients that survived (P = .0003). Cumulative survival was 85% in patients with cluster 1 molecular profiling and 98% in those with cluster 2 molecular profiling. Univariate predictors of mortality in the training cohort included cluster 1 molecular profiling (HR, 7.385; 95% CI, 1.65-33.000; P = .00885), WBC count (HR, 1.145; 95% CI, 1.012-1.295; P = .031), and HbF levels (HR, 0.834; 95% CI, 0.709-0.981; P = .028) (supplemental Table 2). In a multivariable model, adjustment for TRV, WBC, age, and HbF levels did not change the association between cluster 1 molecular profiling and mortality (HR, 6.499; 95% CI, 1.388-30.420; P = .018; Table 2). Kaplan-Meier estimates of survival demonstrated a significant survival difference between patients with cluster 1 molecular profiling compared with those individuals with cluster 2 molecular profiling (P = .002, Figure 2).
Cox regression model in training and testing cohorts
| . | HR (95% CI) . | P value . |
|---|---|---|
| Training cohort | ||
| Cluster 1 | 6.499 (1.388-30.420) | .018 |
| Age (y) | 1.028 (0.979-1.079) | .267 |
| Tricuspid regurgitation velocity (≥2.5 m/s) | 2.239 (0.740-6.776) | .154 |
| WBC count (10−3/mm3) | 1.055 (0.912-1.221) | .472 |
| HbF (%) | 0.827 (0.690-0.991) | .039 |
| Testing cohort | ||
| Cluster 1 | 4.852 (1.269-18.549) | .021 |
| Age (y) | 1.074 (1.023-1.128) | 4.18E-03 |
| Male | 4.056 (1.094-15.042) | .036 |
| . | HR (95% CI) . | P value . |
|---|---|---|
| Training cohort | ||
| Cluster 1 | 6.499 (1.388-30.420) | .018 |
| Age (y) | 1.028 (0.979-1.079) | .267 |
| Tricuspid regurgitation velocity (≥2.5 m/s) | 2.239 (0.740-6.776) | .154 |
| WBC count (10−3/mm3) | 1.055 (0.912-1.221) | .472 |
| HbF (%) | 0.827 (0.690-0.991) | .039 |
| Testing cohort | ||
| Cluster 1 | 4.852 (1.269-18.549) | .021 |
| Age (y) | 1.074 (1.023-1.128) | 4.18E-03 |
| Male | 4.056 (1.094-15.042) | .036 |
Kaplan-Meier survival curves in the training and testing cohorts. When compared with subjects with cluster 2 profiling, subjects with cluster 1 profiling experienced worse overall survival in both the training (A) and testing (B) cohorts. Vertical dashed lines indicate censored observations.
Kaplan-Meier survival curves in the training and testing cohorts. When compared with subjects with cluster 2 profiling, subjects with cluster 1 profiling experienced worse overall survival in both the training (A) and testing (B) cohorts. Vertical dashed lines indicate censored observations.
Validation of the mortality molecular profiling
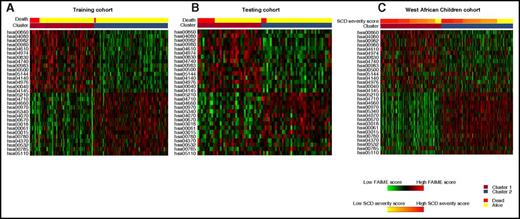
Given the breadth and number of genes within each cluster, a pathway-based classification using FAIME, which computes mechanism scores using rank-weighted gene expression of an individual sample (FAIME28,29 ), was performed to dissect each genome-wide–derived cluster. Using an FDR <1 × 10−5, 30 FAIME-based pathway profiles distinguished the 2 observed clusters in the training cohort. The pathways were heavily represented by increased expression of metabolism of porphyrin and cytochrome 450, among other metabolic pathways, complement and coagulation cascades, bile secretion, and malaria signaling pathways. Significant downregulated pathways included cancer, T-cell receptor signaling, immunodeficiency, and vascular endothelial growth factor (VEGF) signaling pathways (Table 3). Using SVM, the FAIME signature was then validated to predict the 2 transcriptomic clusters and association with overall survival in the testing cohort and the West African cohort (depicted by heat maps in Figure 3).
FAIME pathway signature
| KEGG . | Increased cluster expression . | FDR . | Pathways . |
|---|---|---|---|
| hsa00860 | 1 | 1.62242E-14 | Porphyrin and chlorophyll metabolism |
| hsa04080 | 1 | 3.81206E-12 | Neuroactive ligand-receptor interaction |
| hsa00982 | 1 | 5.25302E-12 | Drug metabolism: cytochrome P450 |
| hsa00980 | 1 | 2.24239E-11 | Cytochrome P450 Metabolism of xenobiotics |
| hsa04610 | 1 | 2.73693E-10 | Complement and coagulation cascades |
| hsa04974 | 1 | 2.75036E-10 | Protein digestion and absorption |
| hsa00830 | 1 | 9.10577E-10 | Retinol metabolism |
| hsa04740 | 1 | 3.45204E-09 | Olfactory transduction |
| hsa00983 | 1 | 1.01799E-08 | Drug metabolism: other enzymes |
| hsa00500 | 1 | 1.1871E-08 | Starch and sucrose metabolism |
| hsa05144 | 1 | 1.23834E-08 | Malaria |
| hsa04140 | 1 | 3.1348E-07 | Regulation of autophagy |
| hsa04976 | 1 | 1.03166E-06 | Bile secretion |
| hsa00040 | 1 | 2.3345E-06 | Pentose and glucuronate interconversions |
| hsa04145 | 1 | 2.81301E-06 | Phagosome |
| hsa05210 | 2 | 1.51002E-12 | Colorectal cancer |
| hsa04710 | 2 | 3.91953E-11 | Circadian rhythm: mammal |
| hsa04660 | 2 | 2.75036E-10 | T-cell receptor signaling pathway |
| hsa00970 | 2 | 1.82174E-08 | Aminoacyl-tRNA biosynthesis |
| hsa05340 | 2 | 1.82174E-08 | Primary immunodeficiency |
| hsa04070 | 2 | 2.32538E-08 | Phosphatidylinositol signaling system |
| hsa00670 | 2 | 3.3263E-08 | One carbon pool by folate |
| hsa03018 | 2 | 1.89728E-07 | RNA degradation |
| hsa00061 | 2 | 2.35402E-07 | Fatty acid biosynthesis |
| hsa03015 | 2 | 3.44253E-07 | mRNA surveillance pathway |
| hsa00780 | 2 | 9.72831E-07 | Biotin metabolism |
| hsa04370 | 2 | 1.69033E-06 | VEGF signaling pathway |
| hsa00532 | 2 | 2.68975E-06 | Glycosaminoglycan biosynthesis chondroitin |
| hsa00785 | 2 | 3.19258E-06 | Lipoic acid metabolism |
| hsa05110 | 2 | 7.04905E-06 | Vibrio cholerae infection |
| KEGG . | Increased cluster expression . | FDR . | Pathways . |
|---|---|---|---|
| hsa00860 | 1 | 1.62242E-14 | Porphyrin and chlorophyll metabolism |
| hsa04080 | 1 | 3.81206E-12 | Neuroactive ligand-receptor interaction |
| hsa00982 | 1 | 5.25302E-12 | Drug metabolism: cytochrome P450 |
| hsa00980 | 1 | 2.24239E-11 | Cytochrome P450 Metabolism of xenobiotics |
| hsa04610 | 1 | 2.73693E-10 | Complement and coagulation cascades |
| hsa04974 | 1 | 2.75036E-10 | Protein digestion and absorption |
| hsa00830 | 1 | 9.10577E-10 | Retinol metabolism |
| hsa04740 | 1 | 3.45204E-09 | Olfactory transduction |
| hsa00983 | 1 | 1.01799E-08 | Drug metabolism: other enzymes |
| hsa00500 | 1 | 1.1871E-08 | Starch and sucrose metabolism |
| hsa05144 | 1 | 1.23834E-08 | Malaria |
| hsa04140 | 1 | 3.1348E-07 | Regulation of autophagy |
| hsa04976 | 1 | 1.03166E-06 | Bile secretion |
| hsa00040 | 1 | 2.3345E-06 | Pentose and glucuronate interconversions |
| hsa04145 | 1 | 2.81301E-06 | Phagosome |
| hsa05210 | 2 | 1.51002E-12 | Colorectal cancer |
| hsa04710 | 2 | 3.91953E-11 | Circadian rhythm: mammal |
| hsa04660 | 2 | 2.75036E-10 | T-cell receptor signaling pathway |
| hsa00970 | 2 | 1.82174E-08 | Aminoacyl-tRNA biosynthesis |
| hsa05340 | 2 | 1.82174E-08 | Primary immunodeficiency |
| hsa04070 | 2 | 2.32538E-08 | Phosphatidylinositol signaling system |
| hsa00670 | 2 | 3.3263E-08 | One carbon pool by folate |
| hsa03018 | 2 | 1.89728E-07 | RNA degradation |
| hsa00061 | 2 | 2.35402E-07 | Fatty acid biosynthesis |
| hsa03015 | 2 | 3.44253E-07 | mRNA surveillance pathway |
| hsa00780 | 2 | 9.72831E-07 | Biotin metabolism |
| hsa04370 | 2 | 1.69033E-06 | VEGF signaling pathway |
| hsa00532 | 2 | 2.68975E-06 | Glycosaminoglycan biosynthesis chondroitin |
| hsa00785 | 2 | 3.19258E-06 | Lipoic acid metabolism |
| hsa05110 | 2 | 7.04905E-06 | Vibrio cholerae infection |
KEGG, Kyoto Encyclopedia of Genes and Genomes; mRNA, messenger RNA; tRNA, transfer RNA.
Heat map depicting FAIME scores of 30 signature pathways stratified by clusters and overall survival. (A) Heat map of scores of all of the differentially regulated pathways between those who survived (yellow, top) and died (red, top) as well as those in clusters 1 (orange, top) and 2 (blue, top) within the training cohort. (B) Heat map of scores from the testing cohort. The panel depicts a heat map of scores of all of the differentially regulated pathways stratified by clusters 1 (orange, top) and 2 (blue, top). (C) SCD severity scores (yellow to red, top) within the West African Children cohort. Red and green in the panels indicate an increase or decrease in gene expression, respectively.
Heat map depicting FAIME scores of 30 signature pathways stratified by clusters and overall survival. (A) Heat map of scores of all of the differentially regulated pathways between those who survived (yellow, top) and died (red, top) as well as those in clusters 1 (orange, top) and 2 (blue, top) within the training cohort. (B) Heat map of scores from the testing cohort. The panel depicts a heat map of scores of all of the differentially regulated pathways stratified by clusters 1 (orange, top) and 2 (blue, top). (C) SCD severity scores (yellow to red, top) within the West African Children cohort. Red and green in the panels indicate an increase or decrease in gene expression, respectively.
During a median follow-up of 5.1 years (95% CI, 4.07-5.38), 16.6% of patients (n = 13) died in the testing cohort. Ten of the patients that died and 20 that survived exhibited cluster 1 molecular profiling, whereas cluster 2 molecular profiling was present in 3 of the patients that died and 38 that survived (P = .031). The cumulative survival was 73% in patients with cluster 1 molecular profiling and 93% in patients with cluster 2 molecular profiling. The higher mortality rates in the testing cohort likely reflect a longer duration of follow-up in the testing cohort. Less likely are that differences in care delivery or unmeasured environmental and biological factors could also contribute to the discrepancy in mortality rates between the cohorts. Univariate predictors of mortality in the testing cohort included cluster 1 molecular profiling (HR, 4.852; 95% CI, 1.269-18.549; P = .021), age (HR, 1.057; 95% CI, 1.016-1.099; P = .00562), and male sex (HR, 4.6; 95% CI, 1.260-16.79; P = .021) (supplemental Table 3). In a multivariable model, adjustment for age and sex did not change the association between cluster 1 molecular profiling and mortality (HR, 4.852; 95% CI, 1.269-18.549; P = .021, Table 2). Similar to the training cohort, Kaplan-Meier estimates of survival demonstrated a significant survival difference between patients with cluster 1 molecular profiling compared with those individuals cluster 1 molecular profiling in the testing cohort (P = .024, Figure 2).
The classifier was next evaluated in children with SCD. With lower mortality rates for children in relation to adults with SCD, severity of illness was examined instead of mortality, defining an “at-risk” population. The definition of disease severity was based on the association of mortality with a published scoring system27 from 3380 SCD patients (children and adults) founded on well-established clinical characteristics (sex, Hb genotype, mean corpuscular volume, and WBC counts). The scores, ranging between 0 (least severe) and 1 (most severe), were predictive of the risk of death within 5 years.27 This risk score was defined for 250 children from West Africa with available whole blood transcriptomic data.26 West African children with SCD that exhibited the FAIME-based pathway signature and cluster 1 profiling demonstrated significantly higher severity scores than those in cluster 2 (P = 9.99 × 10−5; supplemental Figure 2).
To generate a summarized list of transcripts representative of clusters 1 and 2, we overlapped all genes differentially regulated between clusters 1 and 2 in both the training and testing cohorts, resulting in a set of 31 genes (supplemental Table 4). The 31-gene signature was able to differentiate circulating whole blood–derived global transcriptomic cluster profiling in children with varying SCD severity indices (supplemental Figure 2). For further validation, gene expression was evaluated by reverse transcriptase quantitative polymerase chain reaction of 10 of these genes and correlated with microarray-based expression. The fold changes of gene expression determined by reverse transcriptase quantitative polymerase chain reaction highly correlated with those estimated by microarray profiling (Pearson r = 0.99, P = 7.628e-08; supplemental Figure 3).
Risk score combining molecular and clinical data predicts poor prognosis in SCD
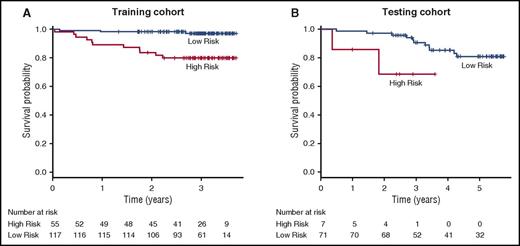
Based on well-established clinical biomarkers for disease severity and mortality in SCD, a risk score was developed integrating these biomarkers with the transcriptomic classifier. This risk score was composed of assigning weights to cluster 1 profiling, along with risk factors including a history of ACS, an elevated TRV (≥2.5 m/s), an elevated WBC count (≥103/μL), and a low Hg level (≤10 g/dL), demonstrated a higher risk for mortality in both the training (HR, 8.268; 95% CI, 2.306-29.640; P = 1.180 × 10−3) and testing (HR, 4.840; 95% CI, 1.004-23.330; P = .049) cohorts when compared with the risk score derived only from the clinical risk parameters (training: HR, 7.527; 95% CI, 1.685-33.630; P = 8.220 × 10−3; testing: HR, 1.888; 95% CI, 0.400-8.916; P = .422) or from cluster 1 data alone (training: HR, 7.385; 95% CI, 1.653-33.000; P = 8.850 × 10−3; testing: HR, 3.957; 95% CI, 1.089-14.380; P = .037) (supplemental Table 5). Kaplan-Meier estimates demonstrated a significant survival difference between patients with “high” composite risk scores (defined as ≥4 points) compared with those patients with “low” composite risk scores (defined as <4 points) in both the training (P = .000105) and testing cohorts (P = .03) when using a combined risk score derived from both clinical risk factors and genomic clustering profile (Figure 4).
Kaplan-Meier survival curves in training and testing cohorts by composite risk score. When compared with subjects with a low composite risk score, subjects with a high composite risk score experienced worse overall survival in both the training (A) and testing (B) cohorts. Vertical dashed lines indicate censored observations.
Kaplan-Meier survival curves in training and testing cohorts by composite risk score. When compared with subjects with a low composite risk score, subjects with a high composite risk score experienced worse overall survival in both the training (A) and testing (B) cohorts. Vertical dashed lines indicate censored observations.
Discussion
We present a validated circulating global transcriptomic profile that predicts overall survival in adults and is associated with previously published markers of disease severity in a West African cohort of children with SCD, independent of the presence of known clinical risk factors. The consistency of the risk expression profiling in heterogeneous patient populations and in both whole blood and PBMCs enhances the feasibility of the application of transcriptome profiling as a biomarker in diverse settings. Furthermore, pathway-based analysis of these transcriptomes clusters to signaling cascades known to be pathophysiologically relevant in SCD.7,11,15,19,26,33-36
Our data suggest that the transcriptome classifier is a biomarker that reflects disease severity as evidenced by abnormalities in important pathways that are driven by HbS polymerization and reflect the systemic nature of the complications of SCD. As such, FAIME analyses provide insights into the potential mechanistic and pathophysiological relevance of molecular clusters downstream of the inciting HgS polymerization event in SCD. Seemingly heterogeneous pathways in porphyrin metabolism, bile secretion, and complement and coagulation cascades along with VEGF signaling and immune-mediated processes have all been shown to be directly relevant to SCD.7,11,15,19,26,33-36 Recently, meta-analysis of 4 published datasets of differentially regulated molecular pathways related to SCD demonstrated near identical pathways to those that are associated with survival in the current work including porphyrin metabolism, complement and coagulation cascades, VEGF signaling, and immune-mediated processes.15 Another group evaluated differentially regulated genes by both RNA-sequencing and conventional gene expression profiling in whole blood from patients with SCD and found identical genes to those found in the current gene signature including BCL2L1, DOCK family candidates, SELENBP1, ALAS2, BSG, FECH, GYPA, and BPGM.16 In fact, nearly all of the unregulated genes in the gene signature (SELENBP1, SLC4A1, EPB42, ALAS2, GYPA, FECH) are associated with erythropoiesis, highlighting the role of hemolysis, red cell/globin synthesis red cell, and globin synthesis. These reports further validate the gene and pathway associations of the current work. Whether the association of these peripherally expressed pathways and genes to outcomes is in part because of their representation as novel and more accurate molecular markers of red cell turnover and hemolysis needs further investigation.
The role of PBMCs and whole blood as biomarkers and as pathophysiologic markers of SCD complications has been documented by several investigators.15,17-19,35,37-39 Gene expression of PBMCs from patients with SCD have exhibited associations with iron homeostasis, inflammation, immunity, and the role or T cells, hypoxic response, and PH.15,19,35 PBMCs comprise lymphocytes and monocytes and, occasionally, nucleated red blood cells. Cheadle et al40 have reported an enrichment in erythroid-specific gene expression in PBMCs associated with disease severity in patients with idiopathic and scleroderma-associated pulmonary arterial hypertension. Similarly, the pathways represented in the current analyses (and some of the genes in our 31-gene signature) could, in part, reflect an enrichment of erythroid-specific gene expression, possibly from nucleated red blood cells, reflecting increased red blood cell turnover from increased SCD severity. Previous work has also shown the role of NF-κB signaling in blood mononuclear cells in regulating endothelial tissue factor expression in sickle transgenic mice with implications for the coagulopathy of SCD.41 The current data further emphasize the association of pathways such as inflammation, T-cell signaling, and coagulation in the development of SCD complications from HgS polymerization.
Patients in cluster 1 demonstrated a greater propensity toward a more severe clinical profile as evidenced by a higher WBC count, a trend toward more severe hemolytic anemia, and history of ACS, all of which are markers of poor outcomes in SCD.8,30-32 Importantly, a composite index integrating these clinical biomarkers with the transcriptomic cluster profiling provided better prognostic information than either of the stratifiers alone. Further, the high-risk transcriptome profile remained an independent predictor of death event after adjustment for these and other covariates. Based on the current data, the differentially regulated FAIME pathways may represent a tool that encompasses these different predictors into a common marker of poor prognosis and death in SCD.
The current work presents the validation of a circulating transcriptomic profile that predicts survival in adults with SCD and further stratifies severity of illness in a West African cohort of children with the disease. Despite the inherent limitations of referral bias, including evaluation in a university-based SCD patient population and the heterogeneous nature of our cohorts, the findings stress the importance of validating this genomic biomarker for mortality in all children with and in community populations of patients with SCD in future studies. Because causes of death were not available on all patients in this work, the findings encourage future characterization of the role of PBMCs, top FAIME pathways, and their functional relevance to the development of the variety of etiologies associated with death in SCD. Assessment of the expression and regulation of these targets in end-organ tissues would complement the profiling completed in this study and provide more insight into pathways of the highest pathophysiological relevance. In spite of these limitations, the usefulness of this novel signature as an independent molecular biomarker of prognosis in SCD still remains and demands its further study as a clinical tool in the care of this patient population.
The data reported in this article have been deposited in the Gene Expression Omnibus database (accession numbers GSE84632 [University of Illinois, training], GSE84633 [Howard University, testing], and GSE84634 [University of Chicago, testing]).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are grateful to Sharon Trevino, Zarema Arbieva, and Nancy Casanova for their assistance in recruiting and follow-up. Samples in this study were processed by the University of Illinois Biorepository supported by the Center for Clinical and Translational Science as well as their Core Genomics Facility.
This study was funded by the National Institutes of Health (NIH), National Heart, Lung, and Blood Institute grants K23HL098454, R01HL111656, and R01 HL127342 (R.F.M.), grant R01 HL136603 (A.A.D.), grant K23HL125984 (S.L.S.), and NIH, National Center for Advancing Translational Sciences grant UL1TR000050; the American Heart Association grant 14CRP18910051 (A.A.D.); and the American Thoracic Society Foundation/Pulmonary Hypertension Association (A.A.D.).
Authorship
Contribution: A.A.D. designed research studies, conducted experiments, acquired and analyzed data, provided reagents, and wrote the manuscript; Z.L. designed research studies, analyzed data, and wrote the manuscript; N.B., M.M.-C., X.Z., and Y.L. analyzed data; S.L.S., S.M.N., T.A., A.R.P., R.M.L., and J.G.N.G. acquired data; B.N.S. conducted experiments and acquired and analyzed data; V.R.G. acquired data and provided reagents; and R.F.M. designed research studies, conducted experiments, acquired and analyzed data, provided reagents, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for S.M.N. is Department of Medicine, University of Pittsburgh, Pittsburgh, PA.
Correspondence: Roberto F. Machado, Section of Pulmonary, Critical Care Medicine, Sleep and Allergy, University of Illinois Chicago, 840 S Wood St, Room 920-N, Clinical Science Building, MC 719, Chicago, IL 60612; e-mail: machador@uic.edu; and Ankit A. Desai, Sarver Heart Center, University of Arizona, 1656 E Mabel St, Room 319, Tucson, AZ 85724; e-mail: adesai@shc.arizona.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal