In this issue of Blood, Desai et al validated the association of transcriptomic profiles in mononuclear cells with poor outcomes in 3 cohorts of patients with sickle cell disease (SCD).1
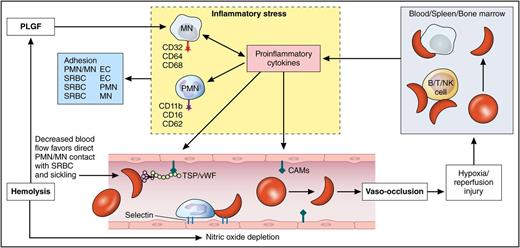
Role of mononuclear cells in the pathophysiology of SCD. Vaso-occlusion and tissue injury lead to activation of the mononuclear cells and the release of proinflammatory cytokines. This leads to further adhesion of sickle red cells and leukocytes to the vascular endothelium perpetuating the vicious cycle of vaso-occlusion and inflammation. CAM, cell adhesion molecule; EC, endothelial cell; MN, monocyte; PLGF, placental growth factor; PMN, polymorphonuclear leukocyte; SRBC, sickle red blood cell; TSP/vWF, thrombospondin/von Willebrand factor. Professional illustration by Patrick Lane, ScEYEnce Studios.
Role of mononuclear cells in the pathophysiology of SCD. Vaso-occlusion and tissue injury lead to activation of the mononuclear cells and the release of proinflammatory cytokines. This leads to further adhesion of sickle red cells and leukocytes to the vascular endothelium perpetuating the vicious cycle of vaso-occlusion and inflammation. CAM, cell adhesion molecule; EC, endothelial cell; MN, monocyte; PLGF, placental growth factor; PMN, polymorphonuclear leukocyte; SRBC, sickle red blood cell; TSP/vWF, thrombospondin/von Willebrand factor. Professional illustration by Patrick Lane, ScEYEnce Studios.
SCD is a life-threatening disease characterized by severe debilitating episodes of pain with ongoing chronic damage to organs leading to premature death. Although this is caused by a single genetic polymorphism, there is a wide heterogeneity in the clinical presentation and severity.2 There is a lack of validated prognostic biomarkers to predict clinical severity in SCD.3 Accurate biomarkers are desperately needed to aid important clinical decisions in the treatment of SCD.
In patients with SCD, abnormal hemoglobin (Hb) polymerizes when deoxygenated, forming the characteristic sickle-shaped erythrocytes.4 Abnormal polymerization along with adhesion of sickle red cells and leukocytes to the vascular endothelium leads to vascular obstruction, tissue ischemia, and reperfusion injury, provoking an intense inflammatory response.5 Although the molecular mechanism is well known, the mechanisms that lead to the clinical manifestations and disease severity are unclear. There is increasing evidence that inflammatory cells unrelated to the β-globin mutation contribute to the pathophysiology of the disease.6 Peripheral blood mononuclear cells (PBMCs) which include lymphocytes (T, B, natural killer [NK] cell) and monocytes play an important role in regulating inflammation. In healthy individuals, they circulate in a resting state continuously monitoring the environment for threats from intruders. When they encounter a pathogen or disease-related state there is loss of regulatory control leading to a robust inflammatory response which is not only directed at the potential pathogen but may also cause deleterious effects and severe symptoms. Inflammatory responses include neutrophil migration, proinflammatory cytokine response, and regulation of cell death (see figure).7
The transcriptome is the set of all messenger RNA in a particular cell or a specific population of cells. Unlike the genome, which is usually fixed for a given cell line, the transcriptome can vary from cell to cell, and with external environmental conditions. The transcriptome reflects the genes that are being actively expressed at any given time in a specific population of cells. Transcriptomic profiling is a powerful screening technology performed by high-throughput analysis often by DNA-based microarray techniques or RNA sequencing.8 Transcriptomic profiles have been described to correlate with inflammatory activated PBMCs in SCD but have not been previously described to predict sickle cell severity.9
Given the significance of inflammation in the pathophysiology of SCD and the role of peripheral mononuclear cells as mediators of inflammation, the authors hypothesized that blood mononuclear transcriptomic profiles might be able to predict the severity of the disease. In a training cohort (172 adult patients with SCD), they used a 31-gene signature to identify 2 clusters of transcriptomic profiles that were able to distinguish 2 groups of patients with different mortality rates. They were able to validate the results in another testing cohort of 78 adult patients with SCD. This cluster-specific classifier was able to predict clinical severity of disease based on a validated severity score in 250 West African children with SCD. A risk score of key clinical markers was then integrated with the transcriptomic classifier. The composite risk score was able to predict mortality better that the clinical markers alone. The genetic results are impressive and validated in heterogeneous cohorts of patients. The cluster-specific transcriptomic profiles of PBMCs and whole blood mononuclear cells, especially when combined with clinical risk factors, seem to be a powerful biomarker for the severity of SCD. The pathway-based analysis of these transcriptomic profiles identified signaling cascades that can be interpreted to have pathophysiologic significance in SCD. These pathways are heterogeneous, and it is not clear whether they are indeed downstream to Hb S polymerization or rather overexpressed as a result of a severe inflammatory response in patients with more severe disease. The findings of Desai et al suggest the need for more work on better understanding how these pathways may be of relevance to various causes of death in SCD. It would be very interesting to further validate the results of this novel genetic biomarker in combination with clinical risk factors in patients with SCD in the rest of the world to see whether similar results are seen across the different populations, including India and the Middle East, to explain some of the genotype-phenotype differences.
Conflict-of-interest disclosure: The author declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal