Key Points
NK cell function testing is less sensitive and no more specific for discriminating genetic HLH compared to perforin and CD107a expression.
Perforin and CD107a testing could augment NK-cell cytotoxicity testing for use in HLH diagnostic criteria.
Abstract
Primary hemophagocytic lymphohistiocytosis (HLH) can be caused by biallelic mutations in PRF1, encoding perforin, or UNC13D, STXBP2, STX11, RAB27A, LYST, and AP3B1, encoding proteins involved in cytotoxic lymphocyte degranulation. Natural killer (NK)–cell cytotoxicity assays can quickly screen for all of these genetic diseases, facilitating treatment, but combining NK-cell perforin expression and CD107a upregulation tests can as well. To determine the relative diagnostic accuracies for each approach, we retrospectively reviewed screening test performance in 1614 patients referred for HLH evaluation. For each test, we generated a receiver operating characteristic (ROC) curve, and calculated area under the curve (AUC) and diagnostic parameters at optimal threshold. We generated an AUC for combining perforin and CD107a tests by creating a logistic regression model and applying model-generated coefficients to patient values. Sensitivities of NK-cell function, perforin mean channel fluorescence (MCF), and CD107a MCF to detect biallelic mutations were 59.5%, 96.6%, and 93.8%, with specificities of 72.0%, 99.5%, and 73%. AUCs for NK-cell cytotoxicity, perforin MCF, CD107a MCF, and combined perforin and CD107a MCFs were 0.690, 0.971, 0.860, and 0.838. Perforin and CD107a tests are more sensitive and no less specific compared with NK cytotoxicity testing for screening for genetic HLH and should be considered for addition to current HLH criteria.
Introduction
Hemophagocytic lymphohistiocytosis (HLH) is a life-threatening disorder characterized by uncontrolled, excessive cytotoxic lymphocyte and macrophage activation, with accompanying massive inflammatory cytokine release.1 The disease can be divided into primary (underlying genetic basis) and secondary forms, which can overlap significantly in their clinical presentation.
Primary HLH can be caused by mutations in several genes, and includes patients with biallelic mutations in PRF1 (encoding perforin) or UNC13D, STXBP2, STX11 (encoding proteins involved in cytotoxic lymphocyte degranulation).2-8 Related hereditary pigmentary disorders, caused by biallelic mutations in RAB27A, LYST, and AP3B1, also impact lymphocyte cytotoxicity and degranulation, and predispose to HLH.9,10 In addition, monoallelic mutations in the X-linked genes SH2D1A and XIAP/BIRC4 can cause primary HLH,11-14 but these mutations typically do not impact global lymphocyte cytotoxicity.14-16 In contrast to primary HLH, secondary HLH tends to occur in the setting of underlying infection, malignancy, rheumatologic or other disease.17 It should be mentioned, however, that in spite of our knowledge of certain genes underlying primary HLH, some patients still have presumed primary forms of HLH, but without a discovered genetic cause.
Despite a somewhat arbitrary distinction between primary and secondary HLH (which, by necessity, is often based on genetic testing of known mutations), this distinction is still useful because it can facilitate implementation of aggressive HLH treatment. Because genetic HLH typically requires hematopoietic stem cell transplant, identifying the genetic forms can also accelerate preparations for eventual allogeneic hematopoietic stem cell transplant.18
The identification of pathogenic mutations in 1 of the genes known to be associated with HLH is the gold standard for diagnosing primary HLH. However, result reporting can take weeks to months. Therefore, screening tests greatly facilitate the diagnostic process, as they can quickly diagnose and categorize patients with primary HLH. The 3 most common currently used screening tests include the chromium release natural killer (NK)–cell cytotoxicity test (also known as the NK cell function test), flow cytometric measurement of cytotoxic lymphocyte perforin expression, and evaluation of NK-cell degranulation using flow cytometric measurement of CD107a upregulation. In addition, for patients suspected of having X-linked lymphoproliferative syndrome type 1 or 2, flow cytometric tests are available to detect the intracellular gene products SAP and XIAP.19
Impaired or absent NK-cell cytotoxicity comprises 1 of the 8 diagnostic criteria that were used for enrollment in the HLH-2004 trial conducted by the Histiocyte Society.20 This test has historically been considered a valid screening tool for patients with genetic defects in cytotoxicity, which includes patients with biallelic mutations in PRF1, UNC13D, STXBP2, STX11, RAB27A, LYST, and AP3B1, but not those with mutations in SH2D1A and XIAP/BIRC4. This test provides a measure of the whole process of NK-cell cytotoxicity, including target cell recognition, effector cell activation, transport and exocytosis of lytic granule contents, and target cell death.21 However, large studies regarding diagnostic accuracy are lacking.
Although not part of the HLH-2004 study diagnostic criteria, the perforin expression and CD107a upregulation tests have been shown to be highly accurate in discriminating patients with primary HLH due to cytotoxicity defects.16,22,23 The former test measures intracytoplasmic perforin in cytotoxic lymphocytes, which has been shown to be reduced in almost all patients with biallelic mutations in PRF1.24 The latter test measures cell surface expression of CD107a on NK cells following exposure to K562 (target) cells. This assay interrogates the lymphocyte granule-mediated cytotoxicity pathway because, at rest, NK cells express CD107a predominantly only within intracytoplasmic granule membranes. Upon exposure to target cells, CD107a-containing intracytoplasmic granule membranes from normal NK cells fuse with the outer NK-cell membrane, resulting in detectable surface CD107a expression.16 Patients with defects in genes involved in the cytotoxic degranulation pathway (which includes those with biallelic mutations in UNC13D, STXBP2, STX11, RAB27A, LYST, and AP3B1, but not patients with mutations in PRF1, SH2D1A, and XIAP/BIRC4 genes) have been shown to exhibit decreased NK-cell surface CD107a upregulation upon exposure to target cells.16,25
Although the diagnostic accuracies of NK-cell perforin expression23 and NK-cell degranulation tests22 for primary HLH have recently been reported, their diagnostic performance has not been compared with the NK cytotoxicity test. Furthermore, diagnostic accuracy of the NK cytotoxicity test with respect to genetic HLH has not been evaluated. We sought to compare the diagnostic performance of the 3 currently most widely used screening immunologic tests for detecting biallelic mutations in any of the known genes affecting global cytotoxic lymphocyte cytotoxicity: PRF1, UNC13D, STXBP2, STX11, RAB27A, LYST, and AP3B1.
Patients, materials, and methods
Samples
Institutional review board approval was obtained. We retrospectively reviewed all clinical samples submitted for HLH gene panel testing at Cincinnati Children’s Hospital Medical Center (CCHMC) from May 2014 (when the panel became clinically available) until June 2016. We also reviewed all samples submitted for individual gene sequencing of PRF1, UNC13D, STXBP2, STX11, RAB27A, LYST, or AP3B1 at CCHMC from June 2013 until June 2016. We reviewed clinical samples tested for any indication, as directed by the ordering physician. We included those samples in our analyses for which any 1 of NK cytotoxicity testing, perforin expression, or CD107a expression was also requested by the ordering physician, and performed by the Diagnostic Immunology Laboratory at CCHMC (Figure 1).
If a patient had multiple results for a given immunologic test, only the first reportable result was included. In total, 1614 patient samples were reviewed. Twelve patients found to have pathologic, disease-causing mutations in genes other than the ones of interest (2 SH2D1A, 8 XIAP/BIRC4, 2 SLC7A) were excluded. Fifteen samples for which NK cytotoxicity testing was performed (15 of 734, 2.0%) were excluded because lytic units were not reportable, due to a nonlinear cytotoxicity response curve, or lack of titration among the 4 effector-to-target dilutions. Twenty-one samples for which perforin expression was performed (21 of 598, 3.5%) were excluded because perforin mean channel fluorescence (MCF) results were not reportable, due to technical performance issues. Four samples for which CD107a expression was performed (4 of 362, 1.1%) needed to be excluded because CD107a MCF results were not reportable due to technical performance issues.
After exclusions, there were 1562 patient samples remaining for analysis. The sample group included 778 female patients (49.8%). The age range for all samples was 0 to 78.22 years, with an average age of 14.04 years and a median age of 8.99 years. There were 719 samples with NK cytotoxicity results, 577 samples with perforin expression and PRF1 sequencing results, 358 samples with CD107a expression and degranulation gene sequencing results, and 276 samples with results for all 3 screening tests. We considered “degranulation gene sequencing” to have been performed if at least UNC13D plus STXBP2 genes were sequenced or, if only 1 of UNC13D, STXBP2, STX11, RAB27A, LYST, or AP3B1 genes was sequenced, if biallelic mutations were detected. Samples for which degranulation gene sequencing was performed may or may not have also had PRF1 sequencing performed.
Patient classification
Patients were classified as “affected” with primary (genetic) HLH if they had biallelic pathogenic mutations in any of the following genes: PRF1, UNC13D, STXBP2, STX11, LYST, RAB27A, or AP3B1. They were classified as “carriers” if they had 1 pathogenic mutation in any of the above-listed genes, and as having “variants of uncertain clinical significance” (VUCS) if they had 1 or more nucleotide changes of uncertain clinical significance. The common A91V variant in the PRF1 gene was included in the VUCS category.
Patients were classified as having normal and abnormal results for each immunologic test based on laboratory-defined normal ranges at our center. A sample’s NK cytotoxicity was considered absent if the lytic unit value was 0, low if the lytic unit value was less than the lower limit of normal of 2.6 but still detectable, and normal if the lytic unit value was ≥2.6. A sample’s NK-cell perforin expression was considered low if perforin MCF was below the lower limit of normal of 98, and normal if the NK-cell perforin MCF was ≥98. A sample’s NK-cell CD107a expression was classified as low if NK-cell CD107a MCF was below the lower limit of normal of 207 but still detectable, and normal if NK-cell CD107a MCF was ≥207. Perforin and CD107a MCF were chosen for analysis because preliminary analyses revealed better accuracy compared with percentage of perforin and CD107a expression, respectively (data not shown).
Genetic testing
DNA was prepared from peripheral blood or other tissue using standard methods. Genetic testing was performed in the Molecular Genetics Laboratory at CCHMC, either by next-generation sequencing (HLH gene panel) or Sanger sequencing of individual genes, also using standard methods. The next-generation–based HLH gene panel at CCHMC offers coverage of >20-fold at every target base, and all bioinformatically determined pathogenic and novel variants and VUCS are confirmed by Sanger sequencing. DNA sequencing is over 99% sensitive for detection of nucleotide base changes, small deletions, and insertions in the regions analyzed. Large deletions/duplication and other complex structural defects are not detected by next-generation sequencing or Sanger sequencing.
Screening assays
NK-cell function, perforin expression, and CD107a upregulation assays were performed by the Diagnostic Immunology Laboratory at CCHMC per standard laboratory protocols. As per protocol, all clinical samples received at the laboratory within 24 hours of being drawn underwent testing, whereas those received beyond 24 hours of being drawn were rejected. Shipping controls were not required by our laboratory. The protocol for perforin expression by flow cytometry has been described in detail previously.17 Details of the NK cytotoxicity and CD107a expression assays are described in the supplemental Methods (available on the Blood Web site).
Statistical analysis
We evaluated the sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) of each screening test (NK-cell cytotoxicity, perforin expression, and CD107a expression) for predicting biallelic mutations, based on the laboratory-generated normal ranges for each test. Violin plots were generated with the vioplot package26 for R.27 For each test, we also performed receiver operating characteristic (ROC) analysis28 to determine the optimal threshold that would identify patients with biallelic mutations with maximum sensitivity and specificity (Youden index).29 Analysis was performed using XLSTAT (Addinsoft, Paris, France).
To assess the diagnostic performance of combining perforin and CD107a expression tests, with and without including the NK-cell cytotoxicity test, we created a model to predict the probability of biallelic HLH mutations. To produce areas under ROC figures, we fit a logistic regression model with a linear and quadratic term for each marker under consideration. The coefficients obtained from the regression model were applied to the values of the marker for each patient to obtain the fitted values. Sensitivity and specificity for the fitted values were calculated and plotted in the ROC figures. Calculations were done in R version 3.2.4 using the Epi package.30
Results
NK cytotoxicity has poor diagnostic accuracy for discriminating genetic HLH
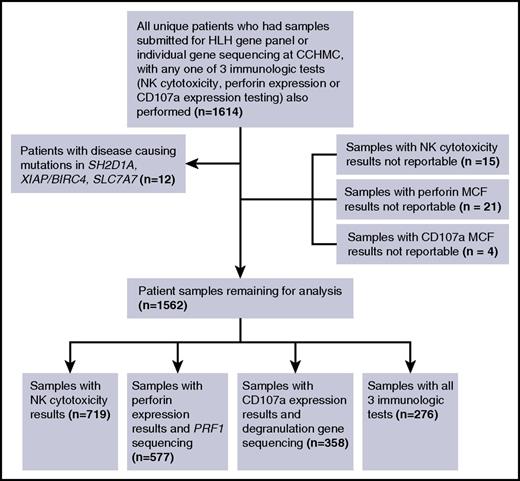
Seventy of 84 patients (83.3%) with biallelic mutations in PRF1 or degranulation genes had low or absent NK lytic units. However, only 271 of 635 patients (42.7%) without biallelic mutations had normal NK lytic units (Figure 2A). Even when excluding carriers or patients with VUCS from the analysis, specificity remained low (43.3%) (Figure 2B). Indeed, a considerable proportion of patients possessing monoallelic mutations (some of whom also possessed VUCS) had low lytic unit values (20 of 38, 52.6%) (Figure 2C). Many patients possessing VUCS (48 of 75, 64%), and with no mutations detected at all (296 of 522, 56.7%), also had low lytic unit values. Whether including or excluding carriers and patients with VUCS from the analysis (Figure 2A-B), PPV of low NK lytic units for genetic HLH was poor at 16.1% and 19.1%, respectively. On the other hand, NPV was excellent (≥94.2%).
Diagnostic accuracy of NK cytotoxicity for detecting biallelic HLH gene mutations. (A) Sensitivity/specificity/PPV/NPV of low NK lytic units to distinguish patients with any biallelic HLH gene mutations (PRF1, UNC13D, STX11, STXBP2, RAB27A, LYST, or AP3B1) from patients with all other sequencing results. (B) Sensitivity/specificity/PPV/NPV of low NK lytic units to distinguish patients with any biallelic HLH mutation from those with normal sequencing results. (C) Distribution of NK lytic unit results among patients with no mutations, 1 or more VUCS, carriers (with or without additional VUCS), and biallelic mutations. (D) ROC curve showing diagnostic accuracy of NK lytic units to distinguish patients with biallelic mutations from patients with all other sequencing results; sensitivity and specificity are shown at the optimal diagnostic cutoff of LU ≤ 0.1.
Diagnostic accuracy of NK cytotoxicity for detecting biallelic HLH gene mutations. (A) Sensitivity/specificity/PPV/NPV of low NK lytic units to distinguish patients with any biallelic HLH gene mutations (PRF1, UNC13D, STX11, STXBP2, RAB27A, LYST, or AP3B1) from patients with all other sequencing results. (B) Sensitivity/specificity/PPV/NPV of low NK lytic units to distinguish patients with any biallelic HLH mutation from those with normal sequencing results. (C) Distribution of NK lytic unit results among patients with no mutations, 1 or more VUCS, carriers (with or without additional VUCS), and biallelic mutations. (D) ROC curve showing diagnostic accuracy of NK lytic units to distinguish patients with biallelic mutations from patients with all other sequencing results; sensitivity and specificity are shown at the optimal diagnostic cutoff of LU ≤ 0.1.
At the optimal diagnostic threshold established by ROC analysis (lytic unit [LU] ≤ 0.1), the sensitivity of NK lytic units to detect patients with biallelic mutations compared with patients with all other sequencing results was 59.5%, and specificity was 72.0%, with an area under the curve (AUC) of 0.690 (Figure 2D). Using the cutoff of LU ≤ 0.1, 148 of 522 samples (28.4%) with normal sequencing results would still have abnormal NK cytotoxicity results. To determine whether the diagnostic performance of the NK cytotoxicity test was more accurate for distinguishing either biallelic PRF1 or biallelic degranulation gene mutations, we applied the same analysis separately to patients with reportable NK lytic unit results and PRF1 sequencing results, and those with reportable NK lytic unit results and degranulation gene sequencing. In both cases, AUCs were still poor, at 0.681 and 0.677, respectively (data not shown).
Measurement of perforin expression has excellent diagnostic accuracy for detecting biallelic PRF1 mutations
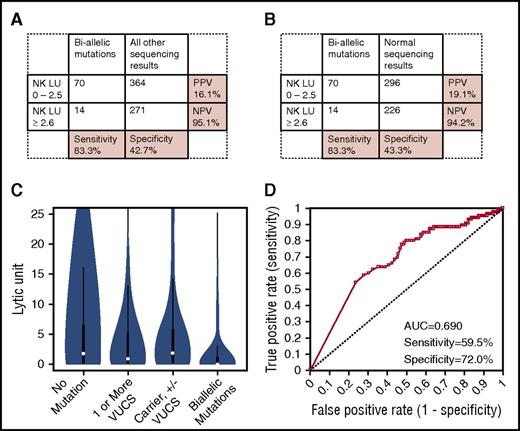
Twenty-eight of 29 patients (96.6%) with biallelic PRF1 mutations had low perforin MCF values, and 456 of 548 patients (83.2%) without biallelic PRF1 mutations had normal perforin MCF values (Figure 3A). Excluding carriers and those with VUCS from the analysis further improved specificity (89.5%) (Figure 3B). Absent perforin MCF expression was highly specific (99.8%) for detecting biallelic pathogenic PRF1 mutations (data not shown). Over half of PRF1 carriers (12 of 23, 52.2%) and those with VUCS (31 of 57, 54.4%) had low perforin results. In contrast, only a minority of those with normal sequencing results (49 of 468, 10.5%) had low perforin results (Figure 3C). Whether including or excluding carriers and patients with VUCS from the analysis, the PPV of laboratory-defined low perforin MCF for biallelic PRF1 mutations was 23.3% or 36.4%, respectively. NPV for perforin MCF was excellent at 99.8%.
Diagnostic accuracy of perforin expression for detecting biallelic PRF1 mutations. (A) Sensitivity/specificity/PPV/NPV of low perforin MCF to distinguish biallelic PRF1 mutations from all other sequencing results. (B) Sensitivity/specificity/PPV/NPV of low perforin MCF to distinguish biallelic PRF1 mutation from patients with normal sequencing results. (C) Distribution of perforin MCF results among patients with no mutations, 1 or more VUCS (including A91V mutations), patients with A91V VUCS, carriers (with or without additional VUCS), and patients with biallelic mutations. (D) ROC curve showing diagnostic accuracy of perforin MCF to distinguish patients with biallelic PRF1 mutations compared with patients with all other sequencing results; sensitivity and specificity are shown at the optimal diagnostic cutoff of perforin MCF ≤ 38.
Diagnostic accuracy of perforin expression for detecting biallelic PRF1 mutations. (A) Sensitivity/specificity/PPV/NPV of low perforin MCF to distinguish biallelic PRF1 mutations from all other sequencing results. (B) Sensitivity/specificity/PPV/NPV of low perforin MCF to distinguish biallelic PRF1 mutation from patients with normal sequencing results. (C) Distribution of perforin MCF results among patients with no mutations, 1 or more VUCS (including A91V mutations), patients with A91V VUCS, carriers (with or without additional VUCS), and patients with biallelic mutations. (D) ROC curve showing diagnostic accuracy of perforin MCF to distinguish patients with biallelic PRF1 mutations compared with patients with all other sequencing results; sensitivity and specificity are shown at the optimal diagnostic cutoff of perforin MCF ≤ 38.
At the optimal diagnostic threshold established by ROC analysis (perforin MCF ≤ 38), sensitivity of perforin MCF to detect patients with biallelic mutations compared with patients with all other genetic results was 96.6%, and specificity was 99.5%, with an AUC of 0.971 (Figure 3D).
CD107a expression has good diagnostic accuracy for detecting biallelic degranulation gene mutations
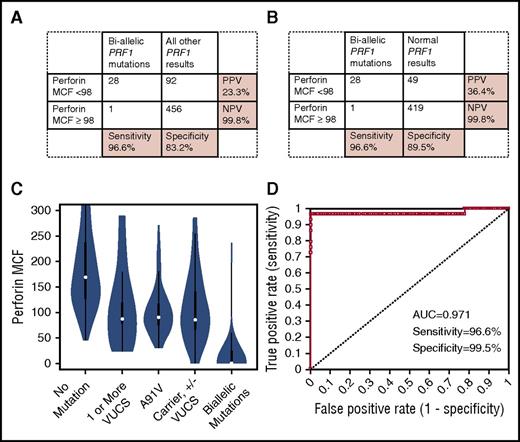
Thirty of 32 patients (93.8%) with biallelic mutations in an HLH-associated degranulation gene had low CD107a MCF (Figure 4A), and 197 of 326 patients (60.4%) without biallelic degranulation gene mutations had normal CD107a MCF. Eliminating carriers or patients possessing VUCS from the analysis did not improve specificity (60.5%) (Figure 4B). In fact, proportions of patients with abnormal degranulation results were distributed fairly evenly across carriers (some of whom possessed additional VUCS [3 of 11, 27.3%]), patients possessing VUCS (8 of 16, 50.0%), and those with normal sequencing results (118 of 299, 39.5%) (Figure 4C). Whether including or excluding carriers, and those possessing VUCS, PPV for low CD107a MCF was low, at 18.9% and 20.3%, respectively. Nevertheless, NPV was excellent (≥98.9%).
Diagnostic accuracy of CD107a expression for detecting biallelic degranulation gene mutations. (A) Sensitivity/specificity/PPV/NPV of low CD107a MCF to distinguish any biallelic degranulation gene mutation (UNC13D, STX11, STXBP2, RAB27A, LYST, or AP3B1) from patients with all other genetic-sequencing results. (B) Sensitivity/specificity/PPV/NPV of low CD107a MCF to distinguish any biallelic degranulation gene mutation from patients with normal genetic results. (C) Distribution of CD107a MCF results among patients with no mutations, 1 or more VUCS, carriers (with or without additional VUCS), and patients with biallelic mutations. (D) ROC curve showing diagnostic accuracy of CD107a MCF to distinguish patients with biallelic degranulation gene mutations compared with patients with all other sequencing results; sensitivity and specificity are shown at the optimal diagnostic cutoff of CD107a MCF ≤ 143.
Diagnostic accuracy of CD107a expression for detecting biallelic degranulation gene mutations. (A) Sensitivity/specificity/PPV/NPV of low CD107a MCF to distinguish any biallelic degranulation gene mutation (UNC13D, STX11, STXBP2, RAB27A, LYST, or AP3B1) from patients with all other genetic-sequencing results. (B) Sensitivity/specificity/PPV/NPV of low CD107a MCF to distinguish any biallelic degranulation gene mutation from patients with normal genetic results. (C) Distribution of CD107a MCF results among patients with no mutations, 1 or more VUCS, carriers (with or without additional VUCS), and patients with biallelic mutations. (D) ROC curve showing diagnostic accuracy of CD107a MCF to distinguish patients with biallelic degranulation gene mutations compared with patients with all other sequencing results; sensitivity and specificity are shown at the optimal diagnostic cutoff of CD107a MCF ≤ 143.
At the optimal diagnostic threshold established by ROC analysis (CD107a MCF ≤ 143), sensitivity of CD107a MCF to detect patients with biallelic mutations compared with patients with all other sequencing results was 93.8%, and specificity was 73%, with an AUC of 0.860 (Figure 4D). Using the cutoff of CD107a MCF ≤ 143, 77 of 299 samples (25.8%) with normal sequencing results would still have abnormal CD107a MCF results.
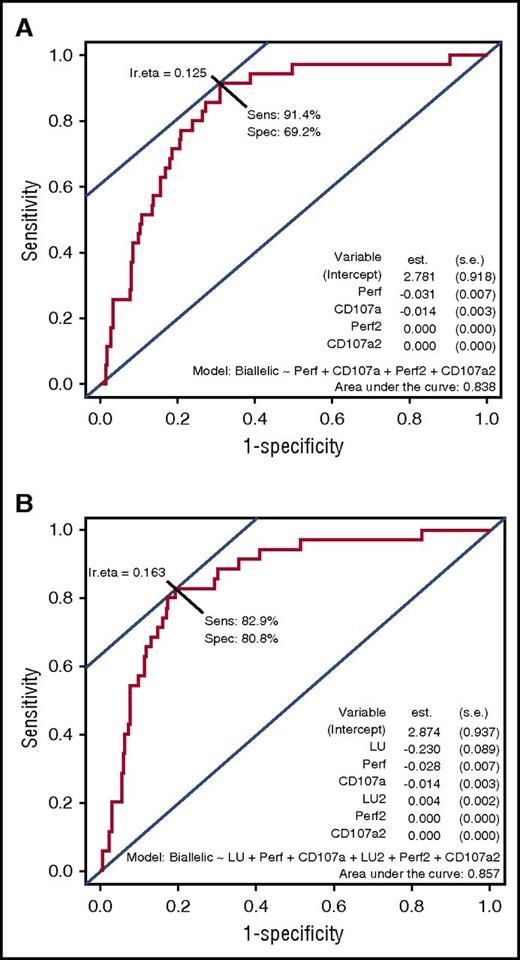
A logistic regression model revealed that combining perforin and CD107a screening, with or without NK function testing, yields good diagnostic accuracy for detecting primary HLH in patients who had all 3 tests performed
In order to minimize referral bias, we last analyzed data from 276 patients who underwent perforin, CD107a, and NK cell function testing. Thirty-four of the 276 patients had biallelic HLH gene mutations. Applying patient values to a logistic regression model and generating an ROC figure, combining CD107a MCF plus perforin MCF produced a sensitivity of 91.4% and specificity of 69.2% with an AUC of 0.838. Adding NK cell function testing to the analysis did not greatly increase the parameters, with sensitivity of 82.9%, specificity of 80.8%, and an AUC of 0.857 (Figure 5).
Logistic regression model predicting probability of biallelic HLH mutations when combining perforin and CD107a expression tests, with or without NK cytotoxicity. ROC display of logistic regression model, showing diagnostic performance of testing with perforin MCF and CD107a MCF together in the model (A) and perforin MCF, CD107a MCF, and NK lytic units together in the model (B) to distinguish patients with biallelic HLH-associated mutations compared with patients with all other genetic results. est., coefficient of the logistic regression model; lr.eta, optimal cut point for the fitted values obtained from the logistic regression model; s.e., standard error for the coefficient; Sens, sensitivity; Spec, specificity.
Logistic regression model predicting probability of biallelic HLH mutations when combining perforin and CD107a expression tests, with or without NK cytotoxicity. ROC display of logistic regression model, showing diagnostic performance of testing with perforin MCF and CD107a MCF together in the model (A) and perforin MCF, CD107a MCF, and NK lytic units together in the model (B) to distinguish patients with biallelic HLH-associated mutations compared with patients with all other genetic results. est., coefficient of the logistic regression model; lr.eta, optimal cut point for the fitted values obtained from the logistic regression model; s.e., standard error for the coefficient; Sens, sensitivity; Spec, specificity.
Discussion
We have reported the diagnostic accuracy of 3 commonly used screening tests for distinguishing patients with biallelic mutations in currently known HLH-associated genes. To our knowledge, this is the first study to assess the performance of the NK cytotoxicity test against genetic sequencing in a large cohort. This study is also the first to compare the diagnostic performance of NK cytotoxicity against perforin expression and CD107a upregulation testing.
In our study, NK-cell perforin MCF and NK-cell CD107a MCF both performed well at distinguishing patients with biallelic HLH-associated mutations, which is consistent with previous reports from our laboratory and others.16,23,31 In contrast, NK-cell cytotoxicity was less sensitive and no more specific for distinguishing patients with biallelic HLH-associated genetic mutations. At the optimal perforin MCF threshold determined by ROC analysis, we found a sensitivity of 96.6%, specificity of 99.5%, and AUC of 0.971. These results are close to those reported by Abdalgani et al (sensitivity, 94%; specificity, 97%; AUC, 0.974).23 We also found that, at an optimal diagnostic threshold, NK-cell CD107a MCF provided excellent sensitivity (93.8%) to distinguish patients with genetic degranulation disorders from patients without genetic degranulation disorders, with a specificity of 73%. A prior study by Bryceson et al showed that abnormal CD107a upregulation below 5% could provide similar sensitivity (96%) for detecting mutations in degranulation genes, albeit with greater specificity (88%).16
Comparing AUCs obtained following ROC analysis, it is clear that NK-cell cytotoxicity (AUC, 0.690) is an inferior test compared with either perforin MCF (0.971) or CD107a MCF (0.860), or the combination of perforin MCF and CD107a MCF (0.838). Although the addition of NK cytotoxicity testing to the combination of perforin and CD107a expression testing could theoretically improve diagnostic accuracy further, the effect was very minimal.
Our findings are clinically relevant because NK cytotoxicity is currently 1 of 8 HLH diagnostic criteria used in the HLH-2004 clinical trial (which have been informally adopted by many clinicians), whereas perforin expression and CD107a expression are not. The chromium release–based NK cytotoxicity test is not an ideal test because it is not widely available, and it is labor intensive. This test utilizes radioactivity (with its attendant obvious safety concerns) and is also impeded in cases of low NK-cell number. Recent difficulty with obtaining 51Cr also demonstrates an additional practical benefit of the flow cytometry tests, which do not require 51Cr. Our finding of poor sensitivity, specificity, and PPV for detecting primary HLH raises additional concerns about its utility. Nevertheless, the NK cytotoxicity test does possess an excellent negative predictive value (>94%), and thus is helpful for ruling out primary HLH. It may also be helpful for clinicians who need to evaluate the functional impact of genetic variants of uncertain significance in HLH-associated genes.
Differentiating primary from acquired forms of HLH may be the single most important function of diagnostic laboratory testing because this distinction often impacts treatment patterns. Our study supports previous smaller studies, which have shown that the 4-hour NK-cell cytotoxicity assay cannot reliably discriminate primary and secondary HLH,32 whereas perforin expression23 and CD107a expression16 can. Furthermore, these latter tests provide an added benefit of helping to guide genetic-sequencing prioritization, and can help with the interpretation of sequence variants of uncertain clinical significance. Flow cytometry–based perforin and CD107a expression testing is available in specialized clinical laboratories in North America and Europe, though not necessarily readily available in other areas.
Our study was limited by its retrospective design and the fact that we limited our analysis to only immunologic laboratory and sequencing results. A further limitation in our study is that, by necessity, we categorized patients with “normal” results for at least 1 allele as not having primary HLH. However, it is possible that at least some of these patients had true primary HLH, as they could have had undetected genetic changes on additional alleles, or mutation(s) in additional as-yet-unknown HLH-causing genes. Because patients with mutations in unknown genes compromising cytotoxic lymphocyte cytotoxicity probably exist, specificities determined for the NK cytotoxicity and CD107a upregulation screening tests could have been falsely lowered in our study. Finally, because our laboratory does not request or require shipping controls, it is also possible that adverse shipping conditioning such as extreme heat or cold could have affected testing results.
Overall, based on our data, we would suggest that perforin and degranulation assays should be preferentially performed to screen patients for primary HLH diseases. Compared with perforin expression and CD107a upregulation, the chromium release NK cytotoxicity test is less sensitive, no more specific, and less informative at a variety of diagnostic values for discriminating patients with primary HLH. We concur with previous recommendations for an HLH diagnostic algorithm that utilizes a combination of flow cytometry tests, including perforin and CD107a expression, as initial screening tests.21 Performing rapid flow cytometry tests for intracellular SAP and XIAP in male patients would additionally identify patients with familial X-linked disorders associated with HLH. We also suggest that perforin expression and degranulation assays should be considered for inclusion in HLH diagnostic criteria used for future studies, to augment the traditional 4-hour chromium release NK-cell cytotoxicity test.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
Salary support for T.S.R. was provided by the Dean of Medicine Education and Research Fund at University of Manitoba.
Authorship
Contribution: T.S.R. analyzed data, interpreted results, and wrote the manuscript; K.Z. supervised the genetic sequencing and interpreted genetic results; A.L. performed statistical analysis; J.J.B. and C.G. developed the assays, supervised sample analyses, and reviewed the manuscript; S.C. analyzed data; and R.A.M. designed the analysis, performed statistical analyses, interpreted results, and edited the manuscript.
Conflict-of-interest disclosure: Revenue generated by the Diagnostic Immunology Laboratory funds a proportion of the salaries for C.G. and J.J.B., and funds the research, development, and validation of clinical tests. The remaining authors declare no competing financial interests.
Correspondence: Rebecca A. Marsh, Division of Bone Marrow Transplantation and Immune Deficiency, Cincinnati Children’s Hospital Medical Center, 3333 Burnet Ave, Cincinnati, OH 45229; e-mail: rebecca.marsh@cchmc.org.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal