Key Points
High expression of PAK4 promotes myeloma cell proliferation through activation of MM antiapoptotic and survival pathways.
Targeting PAK4 with a novel small molecule inhibitor, KPT-9274, has significant impact on MM cell growth and survival.
Abstract
Dysregulated oncogenic serine/threonine kinases play a pathological role in diverse forms of malignancies, including multiple myeloma (MM), and thus represent potential therapeutic targets. Here, we evaluated the biological and functional role of p21-activated kinase 4 (PAK4) and its potential as a new target in MM for clinical applications. PAK4 promoted MM cell growth and survival via activation of MM survival signaling pathways, including the MEK-extracellular signal-regulated kinase pathway. Furthermore, treatment with orally bioavailable PAK4 allosteric modulator (KPT-9274) significantly impacted MM cell growth and survival in a large panel of MM cell lines and primary MM cells alone and in the presence of bone marrow microenvironment. Intriguingly, we have identified FGFR3 as a novel binding partner of PAK4 and observed significant activity of KPT-9274 against t(4;14)-positive MM cells. This set of data supports PAK4 as an oncogene in myeloma and provide the rationale for the clinical evaluation of PAK4 modulator in myeloma.
Introduction
Multiple myeloma (MM) is a hematological malignancy characterized by proliferation of clonal plasma cells in the bone marrow (BM).1 The introduction of novel agents including proteasome inhibitors and immunomodulatory agents alone or in combination has improved outcomes of MM patients.2 However, patients still relapse and ultimately succumb to this disease, providing the impetus to develop novel therapeutic modalities.3 Delineation of signaling pathways mediating MM cell growth, survival, and migration within the BM microenvironment can both enhance our understanding of disease pathogenesis and identify molecular targets for novel MM therapies.
The p21-activated kinase (PAK) family of serine/threonine kinases (STKs) comprises 6 mammalian proteins that are classified into group I (PAK1-3) and group II (PAK4-6) based on structural homology and regulatory function.4 Constitutive activation of PAK1 and 2, positively correlated with increased cell migration potential, has been demonstrated in myeloma cells. We here report high expression of total and phosphorylated (active) PAK4 in the majority of myeloma cell lines, and in all cases of asymptomatic and symptomatic myelomas tested.
As a key downstream effector of the K-Ras pathway and of the ρ-family of GTPases (ρ, Rac, and Cdc42), PAK4 is implicated in a number of intracellular processes, including cytoskeleton reorganization,5 embryonic development,6 as well as cell proliferation, survival, and motility.7 PAK4 is ubiquitously expressed at low levels in many tissues, including BM, and has been found to be overexpressed, genetically amplified, and/or point mutated in several cancer types.8-16 In athymic mice, overexpression or constitutively active form of PAK4 leads to tumor formation, whereas its depletion inhibits tumorigenesis.9 Depletion of PAK4 negatively impacted the activation of NF-қB, extracellular signal-regulated kinase (ERK), and JNK pathways,17 while activating the ATM/Chk1/2/p53 pathway.18 Interestingly, PAK4 may also play a role in gene transcription pathways due to its ability to continuously cycle between the nucleus and the cytoplasm, allowing the modulation of nucleo-cyto trafficking of β-catenin.19
The relative high expression of PAK4 in myeloma and its involvement in major signaling pathways in cancer such as Ras, NF-κB, and Wnt/β-catenin suggests a possible role of PAK4 in myeloma pathogenesis. We here characterized growth and survival activity of PAK4 in myeloma cells and report the therapeutic potential of a novel PAK4 allosteric modulator (PAM).
Material and methods
Cells
Bone marrow mononuclear cells and primary MM cells were isolated using Ficoll-Hypaque density gradient sedimentation from BM aspirates MM patients following informed consent and institutional review board (Dana-Farber Cancer Institute) approval. The human myeloma cell lines (HMMCLs) were cultured in Roswell Park Memorial Institute 1640 medium (RPMI 1640; Mediatech, Herndon, VA) supplemented with 10% fetal bovine serum.
Reagents
Compounds were dissolved in dimethyl sulfoxide (DMSO) unless otherwise stated.
Cell proliferation, viability, and apoptosis assay
MM cell proliferation was measured by (3H)-thymidine (Perkin-Elmer, Boston, MA) incorporation assay, as previously described.20 Cell viability was analyzed by CellTiter Glo (CTG; Promega). Study of caspases activity was performed using caspases 3-7, caspase 8, and caspase 9 Glo assay (Promega). Apoptosis was evaluated by flow cytometric analysis following Annexin V staining. Exvitech automated flow cytometry platform (Vivia Biotech, Madrid, Spain) was used to evaluate activity of KPT-9274 against primary myeloma cells in their microenvironment, as previously described.21 Briefly, BM was diluted with RPMI 1640 to seed 400 to 8000 live cells per well into 96-well plates previously prepared with increasing concentration of KPT-9274 (1 nM-10 µM) and DMSO (up to 0.5%) as vehicle and were incubated for 24 to 72 hours. Then, red cells were lysed with ammonium chloride lysis solution (20 mM KHCO3, 310 mM NH4Cl, 254 µM EDTA). The multiparametric flow cytometry was performed in the ExviTech platform using annexin V and CD138 monoclonal antibody (mAb; Becton Dickinson, San Jose, CA) to identify viable myeloma cells.
Immunoblotting
Western blotting (WB) was performed to delineate expression levels of total protein and phospho-specific isoforms using following antibodies: total PAK4 (Abcam 19007), p-PAK4 (Ser 474) (Santa Cruz Biotechnology), p-CREB (Ser 133) (Cell Signal), pERK (Thr202/Tyr204) (Cell Signal), pAKT (Ser 473) (Cell Signal), pMEK1/2 (Ser217/221) (Cell Signal). Glyceraldehyde-3-phosphatedehydrogenase (GAPDH) was used as loading control (Santa Cruz Biotechnology).
Immunohistochemistry
Immunohistochemical staining of normal and tumor tissue specimen sections of formalin-fixed, paraffin-embedded BM biopsies was prepared and processed for immunohistochemistry to detect PAK4 and p-PAK4 protein expression by using specific antibodies (Abcam, ab62509, and Santa Cruz, sc-135775, respectively).
Antibody-based array screening
The filter arrayed with antibodies (Hypromatrix) was incubated with the lysate of RPMI 8226 cells transfected with green fluorescent protein (GFP)–Pak4 expression plasmid. The antibody-antigen-Pak4 complex was detected by anti-GFP antibody conjugated with horseradish peroxidase.
Inducible gene knockdown
Human TRIPZ PAK4 short hairpin RNA (shRNA) vectors were purchased from Thermo Scientific Bio (Tewksbury, MA). shRNA expression was induced adding 2.5 µg/mL doxycycline to the culturing media. The efficacy of the induction was confirmed by examining the cells microscopically for the presence of TurboRFP and by quantitative polymerase chain reaction (qPCR) after 72 hours of induction. Functional studies were performed as described above.
Clone #23: Clone ID, V3THS_395103
Clone #38: Clone ID, V3THS_335559
Clone #84: Clone ID, V3THS_395101
Clone #87: Clone ID, V3THS_395104
Stable overexpression
LentiORF clone of human PAK4 GFP tagged was purchased from Thermo Scientific Bio (PLOHS_100008678). MM cells were transduced in polybrene media (final concentration, 8 µg/mL) for 8 hours and selected by sorting GFP-positive cells.
Reverse transcriptase-qPCR analysis
Expression of human PAK4 transcript was determined using PAK4 TaqMan Gene Expression Assay, following manufacturer protocols (Applied Biosystems, Foster City, CA). Relative expression was calculated using the comparative δ δ (Ct) method.
In vivo studies
All animal experiments were approved by and conform to the relevant regulatory standards of the Institutional Animal Care and Use Committee at the Dana-Farber Cancer Institute. The in vivo efficacy of KPT-9274 was tested in nude mice subcutaneously injected with either MM1S or OPM2 cell line. Following detection of tumor, mice were treated with either vehicle or KPT-9274 orally. Tumor growth was measured in 2 perpendicular dimensions using a caliper and the following formula: V = (a2 × b)/2, where “a” is the width of the tumor (smaller diameter) and “b” is the length (larger diameter) as previously described.22
Luciferase reporter experiments
The pGL3 Luciferase plasmid containing the NF-қB responsive elements cloned upstream the firefly luciferase reporter gene and the Cignal SRE Reporter Assay Kit (Qiagen) were used to measure NF-қB and ERK activity, respectively. Transfections were performed by electroporation with Neon transfection system (Life Technologies), and Renilla luciferase from pCMV-RL was included to normalize expression of firefly luciferase.
Target identification
Detailed methods can be found in the supplemental Methods, available on the Blood Web site.
Statistical analysis
Data were analyzed using unpaired Student t tests comparing 2 conditions or a 1-way analysis of variance with Bonferroni or Newman-Keuls correction for multiple comparisons using Graphpad software. P < .05 was considered significant. Data are presented as means, and error bars in the figures depict standard deviation.
Results
PAK4 promotes cell growth and survival via activation of MEK/ERK pathway
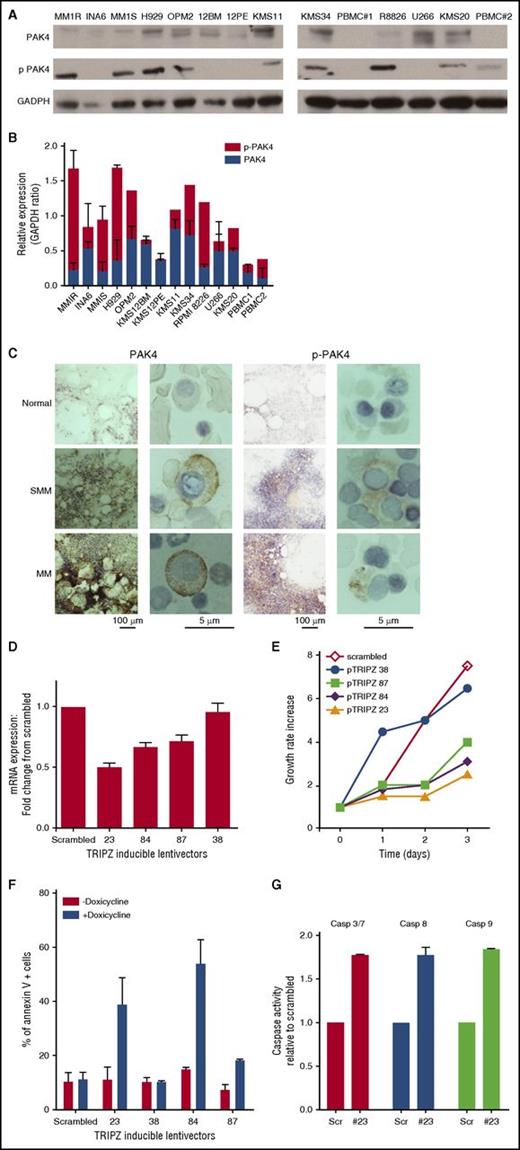
We analyzed expression of PAK4 and phosphorylated (p)-PAK4 Ser474 in different MM cell lines and peripheral blood mononuclear cells (PBMCs) by WB analysis. As seen in Figure 1A-B, PAK4 is highly phopshorylated in the majority of MM cell lines, but not in PBMCs from healthy donors. Protein expression correlated with messenger RNA (mRNA) expression in MM cell lines, as evaluated by RNA-seq (data not shown). We confirmed high expression of PAK4 and (p)-PAK4 in CD138+ MM cells from patients with SMM (n = 3) and MM (n = 9), whereas normal plasma cells were negative for both pPAK4 and total PAK4 (Figure 1C; supplemental Figure 1).
PAK4 expression affects growth and survival in MM. (A) Protein lysates from a panel of MM cell lines were analyzed for PAK4 and p-PAK4 expression by WB. GAPDH was used as loading control. One representative experiment of 2 is shown. (B) PAK4 and p-PAK4 relative expression levels. Data reported represent mean of 3 independent experiments. (C) Representative images of PAK4 and p-PAK4 immunocytochemistry stain in BM from normal, SMM, and symptomatic MM individuals. Scale bars: 100 μm and 5 μm. (D) Genetic depletion of PAK4 was achieved using 4 different tetracycline-inducible pTRIPz-Turbo-RFP vectors (Thermo Scientific, Pittsburgh, PA) containing the target sequence or scrambled control. Transfected MM1S cells were plated in growth medium in the absence or presence of 2.5 μg/mL doxycycline. qPCR analysis (right panel) was performed at day 3, confirming decreased PAK4 mRNA levels in cells expressing inducible PAK4 shRNAs compared with scrambled cells. (E) Cellular proliferation was evaluated by (3H)-thymidine uptake and presented as growth rate increase compared with t = 0. In medium containing doxycycline, reduced expression of PAK4 is accompanied by a reduction of cell growth rate compared with control cells. (F) Apoptosis was evaluated after 3 days of induction with 2.5 μg/mL doxycycline, using Annexin V and propidium iodide (PI) staining followed by flow cytometry acquisition and analysis. (G) Caspases activation was evaluated after 3 days of induction with 2.5 μg/mL doxycycline by luminescence assay.
PAK4 expression affects growth and survival in MM. (A) Protein lysates from a panel of MM cell lines were analyzed for PAK4 and p-PAK4 expression by WB. GAPDH was used as loading control. One representative experiment of 2 is shown. (B) PAK4 and p-PAK4 relative expression levels. Data reported represent mean of 3 independent experiments. (C) Representative images of PAK4 and p-PAK4 immunocytochemistry stain in BM from normal, SMM, and symptomatic MM individuals. Scale bars: 100 μm and 5 μm. (D) Genetic depletion of PAK4 was achieved using 4 different tetracycline-inducible pTRIPz-Turbo-RFP vectors (Thermo Scientific, Pittsburgh, PA) containing the target sequence or scrambled control. Transfected MM1S cells were plated in growth medium in the absence or presence of 2.5 μg/mL doxycycline. qPCR analysis (right panel) was performed at day 3, confirming decreased PAK4 mRNA levels in cells expressing inducible PAK4 shRNAs compared with scrambled cells. (E) Cellular proliferation was evaluated by (3H)-thymidine uptake and presented as growth rate increase compared with t = 0. In medium containing doxycycline, reduced expression of PAK4 is accompanied by a reduction of cell growth rate compared with control cells. (F) Apoptosis was evaluated after 3 days of induction with 2.5 μg/mL doxycycline, using Annexin V and propidium iodide (PI) staining followed by flow cytometry acquisition and analysis. (G) Caspases activation was evaluated after 3 days of induction with 2.5 μg/mL doxycycline by luminescence assay.
To determine the role of PAK4 in MM growth and survival, we suppressed its expression using an inducible knockdown system and observed significant inhibition of MM cell proliferation (Figure 1D-E) and induction of a robust apoptotic response proportional to the reduction in PAK4 levels (Figure 1F). The cells with the most significant reduction in PAK4 mRNA levels (pTRIPZ numbers 23 and 84) showed the greatest induction of apoptosis (39% and 45% of annexin V positive cells, respectively) compared with scrambled or untreated cells (≈10% of annexin V–positive cells). Activation of caspase 3, -8, and -9 was also observed in PAK4-depleted cells (Figure 1G). Importantly, the growth inhibitory effect of PAK4 silencing was confirmed in 2 additional MM cell lines using CRISPR knockdown studies (supplemental Figure 2A). To test whether inducible depletion of PAK4 in MM cells might affect their ability to form tumors in vivo, we treated mice with a doxycycline diet to induce PAK4 knockdown. PAK4 depletion after tumor development significantly inhibited MM tumor growth (supplemental Figure 2B).
Conversely, ectopic expression of PAK4 increased the proliferation rate in the RPMI 8226 MM cell line in vitro (Figure 2A-B). Importantly, overexpression of PAK4 increased the tumorigenic capacity of this cell line in vivo in a xenograft mouse model of human MM: all mice injected with PAK4 overexpressing cells developed tumors at an earlier time point than those receiving control cells, and tumors grew significantly more rapidly (Figure 2C). The growth-promoting effect of PAK4 overexpression was confirmed in 4 additional MM cell lines with intermediate to low levels of either PAK4 or p-PAK4 (Figure 2D-E).
PAK4 triggers MEK/ERK pathway activation in MM cells. (A) WB analysis of PAK4 expression in RPMI 8226 cells overexpressing PAK4. (B) Effect of PAK4 overexpression in RPMI 8226 cells was evaluated over time by (3H)-thymidine uptake and presented as fold change increase compared with day 1. (C) In vivo evaluation of the effects of PAK4 overexpression on MM cells. Growth curve assesses tumor size after injection of an equal number of PAK4 overexpression or empty vector cells subcutaneously into the right posterior flank region of severe combined immunodeficiency mice. Data are shown as the mean values ± standard deviation. (D) PAK4 mRNA levels were evaluated in control and PAK4-overexpressing cells by qPCR analysis. Data are presented as fold change increase from corresponding control cells. (E) Effect of PAK4 overexpression in H929, MM1S, INA6, and KMS12BM MM cell lines was evaluated over time by (3H)-thymidine uptake and presented as fold change increase compared with day 1. (F) WBs showing inhibition of indicated signaling proteins in H929 and RPMI 8226 myeloma cells ectopically expressing PAK4, compared with respective control. GAPDH was used as loading control. One representative blot of 2 is shown. (G) Control and ectopically expressing PAK4 cells were treated with and without U0126 (10 µM) or LY29004 (10 µM) for 48 hours. Cell proliferation was assessed by (3H)-thymidine uptake and presented as count per minute (CPM). (H) MM1S cells transfected with inducible tetracycline-inducible pTRIPz-Turbo-RFP vector #23 or control were treated with 2.5 μg/mL doxycycline for 3 consecutive days. WB analysis was performed using indicated mAbs. *P < .05; **P < .005; ***P < .0005. CNT, control; NS, not significant; WT, wild type.
PAK4 triggers MEK/ERK pathway activation in MM cells. (A) WB analysis of PAK4 expression in RPMI 8226 cells overexpressing PAK4. (B) Effect of PAK4 overexpression in RPMI 8226 cells was evaluated over time by (3H)-thymidine uptake and presented as fold change increase compared with day 1. (C) In vivo evaluation of the effects of PAK4 overexpression on MM cells. Growth curve assesses tumor size after injection of an equal number of PAK4 overexpression or empty vector cells subcutaneously into the right posterior flank region of severe combined immunodeficiency mice. Data are shown as the mean values ± standard deviation. (D) PAK4 mRNA levels were evaluated in control and PAK4-overexpressing cells by qPCR analysis. Data are presented as fold change increase from corresponding control cells. (E) Effect of PAK4 overexpression in H929, MM1S, INA6, and KMS12BM MM cell lines was evaluated over time by (3H)-thymidine uptake and presented as fold change increase compared with day 1. (F) WBs showing inhibition of indicated signaling proteins in H929 and RPMI 8226 myeloma cells ectopically expressing PAK4, compared with respective control. GAPDH was used as loading control. One representative blot of 2 is shown. (G) Control and ectopically expressing PAK4 cells were treated with and without U0126 (10 µM) or LY29004 (10 µM) for 48 hours. Cell proliferation was assessed by (3H)-thymidine uptake and presented as count per minute (CPM). (H) MM1S cells transfected with inducible tetracycline-inducible pTRIPz-Turbo-RFP vector #23 or control were treated with 2.5 μg/mL doxycycline for 3 consecutive days. WB analysis was performed using indicated mAbs. *P < .05; **P < .005; ***P < .0005. CNT, control; NS, not significant; WT, wild type.
Previous studies have shown an association between the PAKs and the RAF/MEK/ERK and PI3K/AKT signaling pathways. To assess the mechanism by which PAK4 contributes to MM cell proliferation, we examined the effect of ectopic PAK4 overexpression or RNA interference–mediated knockdown on RAF/MEK/ERK and PI3K/AKT signaling pathways.23 PAK4 overexpression was associated with activation of NF-κB and MEK-ERK pathways and related induction of CREB (Figure 2F). Interestingly, we have observed a pleiotropic effect on AKT signaling pathway, with a significant activation observed in the H929 cells upon PAK4 overexpression. However, treatment of cells with MEK1 inhibitor U0126, but not the PI3K inhibitor LY294002, significantly reduced PAK4-induced MM growth (Figure 2G), suggesting a significant role of the MEK-ERK pathway in the oncogenic potential of PAK4 in MM. WB analysis confirmed significant impact of PAK4 on the MEK-ERK signaling pathway in response to inducible PAK4 depletion (Figure 2H). Finally, although we observed a significant impact on cytoskeleton-related proteins such as LIMK1 and vinculin in response to PAK4 overexpression and/or inhibition, adhesion and migration processes in MM cells were not perturbed by PAK4 dysregulation (data not shown).
From these observations, we conclude that PAK4 promotes MM cell proliferation both in vitro and in vivo via activation of MEK/ERK survival pathway, representing a potential new therapeutic target for MM.
Structural and functional characterization of novel PAMs
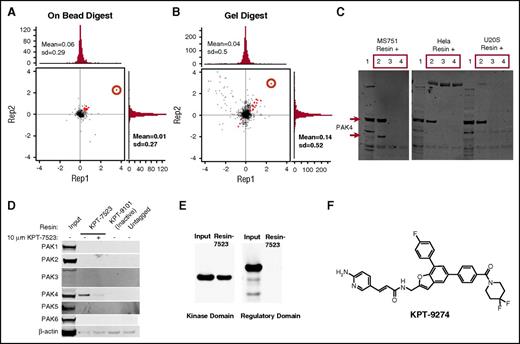
With a significant impact of PAK4 on MM cell growth, we have developed a novel class of orally bioavailable PAMs. Quantitative proteomics methods based on stable isotope labeling of amino acids in cells24 revealed PAK4 (red circle) as the major target of KPT-7523 in the MS-751 cells (3-(4,5-dimethylthiazol-2-yl)-2,5-dimethyltetrazolium bromide; 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide assay 50% inhibitory concentration [IC50] = 30 nM) with minor targets identified as red dots in the graph (Figure 3A-B; Supplemental Table 1). Although KPT-7523 has a Michael acceptor, the analysis of the entire primary sequence by the mass spectrometry did not reveal any evidence of covalent binding of KPT-7523 to PAK4. We also confirmed binding of PAK4 to KPT-7523 resin in an additional 3 cell lines, MS-751, HeLa, and U-2 OS (Figure 3C). Although all 6 PAK family members are structurally similar and group II PAK members have an even greater functional similarity, KPT-7523 binds specifically to PAK4, with only a weak affinity to PAK5 (purified protein; Figure 3D and data not shown).
Discovery and characterization of PAMs. MS-751 cells were labeled with either heavy or light amino acids, lysed, and incubated with ×50 free KPT-7523 or DMSO, respectively, for 2 hours. The lysates were then incubated with KPT-7523 resin overnight. Samples were combined and washed, and then either digested on beads or run on sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), separated by molecular weight ranges, and then digested. Proteins were quantified by mass spectroscopy and analyzed. Both the (A) on-bead digestion and (B) in-gel digestion revealed PAK4 (red circle) as the major target of KPT-7523, with minor targets identified as red dots. (C) Cell lysates from MS-751, HeLa, and U2OS cells were treated with ×50 free KPT-7523, free KPT-7523–polyethylene glycol, or DMSO for 2 hours. The lysates were then passed over KPT-7523 resin overnight at 4°C. The resin was washed with buffer and run on SDS-PAGE for WB with PAK4. PAK4 specifically bound to KPT-7523 resin in all 3 cell lines tested. (D) MDA-MB-231 cells were treated with either DMSO or 10 µM KPT-7523 for 48 hours and then collected by lysing. The lysates were incubated with either KPT-7523 resin, KPT-9101 resin (inactive compound), or untagged resin overnight at 4°C. Resin was washed and run on SDS-PAGE and probed for PAK1-6. KPT-7523 resin bound specifically to PAK4. KPT-9101 and untagged resin did not bind to PAK4. (E) PAK4 regulatory (1-290 aa) and kinase (291-591 aa) domains were produced and purified from E coli. The purified protein was incubated with KPT-7523 resin overnight, washed, and then run on SDS-PAGE. PAK4 antibody measured specific binding of PAK4 kinase domain to KPT-7523. (F) Structure of the clinical candidate KPT-9274.
Discovery and characterization of PAMs. MS-751 cells were labeled with either heavy or light amino acids, lysed, and incubated with ×50 free KPT-7523 or DMSO, respectively, for 2 hours. The lysates were then incubated with KPT-7523 resin overnight. Samples were combined and washed, and then either digested on beads or run on sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), separated by molecular weight ranges, and then digested. Proteins were quantified by mass spectroscopy and analyzed. Both the (A) on-bead digestion and (B) in-gel digestion revealed PAK4 (red circle) as the major target of KPT-7523, with minor targets identified as red dots. (C) Cell lysates from MS-751, HeLa, and U2OS cells were treated with ×50 free KPT-7523, free KPT-7523–polyethylene glycol, or DMSO for 2 hours. The lysates were then passed over KPT-7523 resin overnight at 4°C. The resin was washed with buffer and run on SDS-PAGE for WB with PAK4. PAK4 specifically bound to KPT-7523 resin in all 3 cell lines tested. (D) MDA-MB-231 cells were treated with either DMSO or 10 µM KPT-7523 for 48 hours and then collected by lysing. The lysates were incubated with either KPT-7523 resin, KPT-9101 resin (inactive compound), or untagged resin overnight at 4°C. Resin was washed and run on SDS-PAGE and probed for PAK1-6. KPT-7523 resin bound specifically to PAK4. KPT-9101 and untagged resin did not bind to PAK4. (E) PAK4 regulatory (1-290 aa) and kinase (291-591 aa) domains were produced and purified from E coli. The purified protein was incubated with KPT-7523 resin overnight, washed, and then run on SDS-PAGE. PAK4 antibody measured specific binding of PAK4 kinase domain to KPT-7523. (F) Structure of the clinical candidate KPT-9274.
In order to identify the binding site of KPT-7523, PAK4 regulatory (1-290 amino acids [aa]) and kinase (291-591 aa) domains were produced and purified from Escherichia coli. Immunoprecipitation of the purified peptides with KPT-7523 revealed specific binding only to the PAK4 kinase domain (C-terminal; Figure 3E). In addition, immunoprecipitation in HEK293 cells transfected with FLAG-tagged full-length PAK4, PAK4 kinase domain, or PAK4 regulatory domain showed that KPT-7523 bound preferentially to full-length and KD-PAK4 but not to regulatory domain (supplemental Figure 3A), confirming the results obtained with purified proteins. Therefore, KPT-7523 binds specifically to the PAK4 kinase domain. We further confirmed binding of purified kinase domain or full-length PAK4 to KPT-7523 using isothermal calorimetry and surface plasmon resonance (supplemental Figure 3B-C). The free energy, enthalpy, and entropy suggest a favorable binding.
Structurally similar analogs of KPT-6604, KPT-7189, KPT-8752, and KPT-9274 were next tested against a panel of MM cell lines and PBMCs from healthy donors for their effect on growth and survival. As shown in supplemental Figure 4A for the OPM2 MM cell line, chosen as representative of a panel of 10 cell lines tested, all the compounds decreased MM cell growth and survival in a time- and dose-dependent manner while sparing healthy donor PBMCs, indicating a favorable therapeutic index. To confirm specificity of the effect, we also tested an inactive compound (KPT-9101) and observed no significant effect on both MM growth and survival (supplemental Figure 4B). Based on optimized properties of Absorption, Distribution, Metabolism, and Excretion/Pharmacokinetics, compound KPT-9274 (Figure 3F) was selected for additional preclinical studies as well as for human clinical trials (see clinicaltrials.gov; NCT02702492).
KPT-9274 induces MM cell death in vitro and in vivo, suppressing BM microenvironment-mediated effects
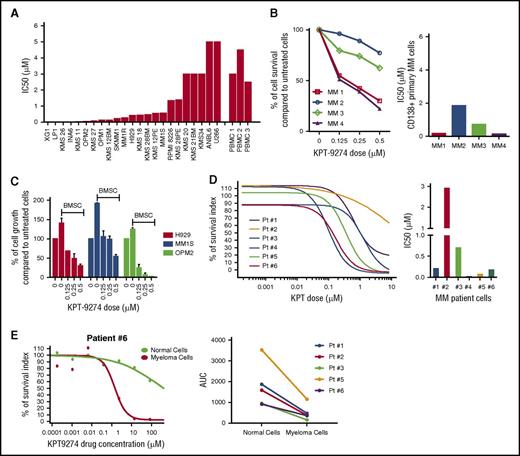
In order to establish the translational potential of targeting PAK4 in myeloma and the clinical applicability of KPT-9274, we evaluated the sensitivity of KPT-9274 in a large panel of human myeloma cell lines representing the different translocation-associated subgroups. Both myeloma cell survival and growth were significantly affected upon treatment with KPT-9274 in a time- and dose-dependent manner in MM cell lines (supplemental Figure 4C-D and data not shown). Importantly, IC50 analysis at 48 hours after treatment showed a greater sensitivity to KPT-9274 in 18 of 23 myeloma cell lines compared with PBMC from healthy donors (Figure 4A). We confirmed significant antimyeloma effect of KPT-9274 in CD138-positive myeloma cells from patients (N = 4) (Figure 4B). Importantly, exogenous PAK4 expression partly rescued the impact of KPT-9274 on MM cell growth, whereas PAK4 depletion by inducible KD did not potentiate KPT-9274 activity against MM cell proliferation (supplemental Figure 4E-F).
KPT-9274 inhibits MM cell growth and survival and overcomes promoting effect of the BM milieu. (A) A panel of 23 HMMCLs was treated with different doses of KPT-9274 for 48 hours, and cell survival was assessed by CTG. IC50 analysis was performed with GraphPad software. (B) CD138+ MM cells from 4 MM patients were cultured in the presence of different concentrations of KPT-9274 for 48 hours. Cell viability was assessed by CTG and expressed as percent change from untreated cells (left panel). IC50 analysis is also shown (right panel). (C) H929, MM1S, and OPM2 cells were cultured with and without BMSC, and in the presence of different doses of KPT-9264 for 48 hours. Cell proliferation was assessed by (3H)-thymidine uptake assay and presented as percent change from untreated cells cultured in the absence of BMSC. (D) BM from 6 myeloma patients was diluted with RPMI to seed 400 to 8000 live cells per well into 96-well plates previously prepared with increasing concentration of KPT-9274 (1 nM-10 µM) and DMSO (up to 0.5%) as vehicle and were incubated for 24 to 72 hours. After red cell lysis, cells were stained with annexin V and CD138 mAb to identify viable myeloma cells. Viable cell number was transformed to percent inhibition relative to vehicle control. Dose-response curves were fitted to nonlinear regression analysis (left panel). IC50 analysis was performed using GraphPad analysis software (right panel). (E) Left panel shows dose-response curve fitted to nonlinear regression analysis of both malignant plasma cells and normal BM cells of a representative case (patient 6). Right panel displays dose/response effect in malignant and normal BM cells of 5 myeloma patients. Data are shown as area under the curve (AUC).
KPT-9274 inhibits MM cell growth and survival and overcomes promoting effect of the BM milieu. (A) A panel of 23 HMMCLs was treated with different doses of KPT-9274 for 48 hours, and cell survival was assessed by CTG. IC50 analysis was performed with GraphPad software. (B) CD138+ MM cells from 4 MM patients were cultured in the presence of different concentrations of KPT-9274 for 48 hours. Cell viability was assessed by CTG and expressed as percent change from untreated cells (left panel). IC50 analysis is also shown (right panel). (C) H929, MM1S, and OPM2 cells were cultured with and without BMSC, and in the presence of different doses of KPT-9264 for 48 hours. Cell proliferation was assessed by (3H)-thymidine uptake assay and presented as percent change from untreated cells cultured in the absence of BMSC. (D) BM from 6 myeloma patients was diluted with RPMI to seed 400 to 8000 live cells per well into 96-well plates previously prepared with increasing concentration of KPT-9274 (1 nM-10 µM) and DMSO (up to 0.5%) as vehicle and were incubated for 24 to 72 hours. After red cell lysis, cells were stained with annexin V and CD138 mAb to identify viable myeloma cells. Viable cell number was transformed to percent inhibition relative to vehicle control. Dose-response curves were fitted to nonlinear regression analysis (left panel). IC50 analysis was performed using GraphPad analysis software (right panel). (E) Left panel shows dose-response curve fitted to nonlinear regression analysis of both malignant plasma cells and normal BM cells of a representative case (patient 6). Right panel displays dose/response effect in malignant and normal BM cells of 5 myeloma patients. Data are shown as area under the curve (AUC).
We next examined the antimyeloma effect of KPT-9274 in the context of the myeloma BM milieu. Treatment with KPT-9274 suppressed MM–bone marrow stromal cells (BMSC) interaction-mediated growth of MM cells (Figure 4C), yet had no effect on the viability of BMSCs (data not shown). Importantly, we evaluated the effect of KPT-9274 in primary myeloma cells from 6 patients cultured in the presence of their respective BM microenvironment using an automated flow cytometry platform for functional drug screening. We observed a dose- and time-dependent effect of KPT-9274 against myeloma cells, whereas normal BM cells derived from the same patient resulted in resistance to the treatment (Figure 4D-E and data not shown).
Interestingly, IC50 analysis showed a greater sensitivity to KPT-9274 in MM cell lines with t(4:14) or harboring FGFR3 gene mutations, such as KMS11 and OPM2 cells (Figure 5A; supplemental Figure 5), and in CD138-positive cells from a t4:14 MM patient with FGFR3/IGH fusion (IC50 = 100 nM) (Figure 5B). KPT-9274 treatment had no significant effect on CD138-negative cells from the same patient. This superior sensitivity to KPT-9274 in t(4:14) myeloma cells was also observed in vivo in mice injected with the t(4:14) FGFR3-mutated OPM2 MM cell line, compared with humanized murine xenograft model of myeloma in which nude mice were subcutaneously injected with the MM1S MM cell line (Figure 5C-D).
Targeting PAK4 by KPT-9274 induces significant cell death in FGFR3-expressing, t(4:14)-positive HMMCLs. (A) Public data from Jonathan Keats’ laboratory (keatslab.org) were used to evaluate the presence of cytogenetic abnormalities, FGFR3 mutations, and FGFR3 expression level in the 23 MM cell lines tested for KPT-9274 sensitivity. Two groups were established based on the presence versus absence of t(4;14), FGFR3 mutation, and high level of FGFR3 expression (superior to the median calculated in the 23 MMCL). Fourteen MMCL were at least positive for t(4;14), high FGFR3 expression, or presence of FGFR3 mutation, whereas 9 MMCL were negative for all. We next compared the KPT-9274 IC50 average between the 2 groups using an unpaired Student t test and observed that MMCL with high FGFR3 and/or FGFR3 mutation and/or t(4;14) were significantly more sensitive (P = .0324). (B) CD138-positive and -negative cells from a t(4;14) myeloma patient with FGFR3/IGH fusion were treated with different concentrations of KPT-9274 for 48 hours. Cell viability was assessed by CTG and presented as percent of viable cells compared with control. IC50 analysis was performed using GraphPad software. (C) Nude mice were subcutaneously inoculated with MM1S (left panel) or OPM2 (right panel) MM cell lines. Treatment started following detection of tumor (∼2 weeks from cell injection). Mice were treated with either 100 mg/kg of KPT-9274 or vehicle orally once per day, 5 d/wk. Tumors were measured in 2 perpendicular dimensions by caliper. (D) Comparison of tumor volume in control and treated mice at day 13 after initial assessment of tumor appearance and start of treatment in MM1S and OPM2 injected mice respectively. (E) A nitrocellulose filter arrayed with antibodies against 400 signal transduction proteins was blotted with lysates of RPMI 8226 cells transfected with GFP-tagged PAK4. The multiprotein complexes were detected by immunoblotting with a horseradish peroxidase–conjugated anti-GFP antibody and visualized by chemiluminescence. (F) Nitrocellulose filters immobilized with 24 antibodies were incubated with whole cell lysates from cells untreated or treated with KPT-9274. Data are presented as signal intensity. TR, treated.
Targeting PAK4 by KPT-9274 induces significant cell death in FGFR3-expressing, t(4:14)-positive HMMCLs. (A) Public data from Jonathan Keats’ laboratory (keatslab.org) were used to evaluate the presence of cytogenetic abnormalities, FGFR3 mutations, and FGFR3 expression level in the 23 MM cell lines tested for KPT-9274 sensitivity. Two groups were established based on the presence versus absence of t(4;14), FGFR3 mutation, and high level of FGFR3 expression (superior to the median calculated in the 23 MMCL). Fourteen MMCL were at least positive for t(4;14), high FGFR3 expression, or presence of FGFR3 mutation, whereas 9 MMCL were negative for all. We next compared the KPT-9274 IC50 average between the 2 groups using an unpaired Student t test and observed that MMCL with high FGFR3 and/or FGFR3 mutation and/or t(4;14) were significantly more sensitive (P = .0324). (B) CD138-positive and -negative cells from a t(4;14) myeloma patient with FGFR3/IGH fusion were treated with different concentrations of KPT-9274 for 48 hours. Cell viability was assessed by CTG and presented as percent of viable cells compared with control. IC50 analysis was performed using GraphPad software. (C) Nude mice were subcutaneously inoculated with MM1S (left panel) or OPM2 (right panel) MM cell lines. Treatment started following detection of tumor (∼2 weeks from cell injection). Mice were treated with either 100 mg/kg of KPT-9274 or vehicle orally once per day, 5 d/wk. Tumors were measured in 2 perpendicular dimensions by caliper. (D) Comparison of tumor volume in control and treated mice at day 13 after initial assessment of tumor appearance and start of treatment in MM1S and OPM2 injected mice respectively. (E) A nitrocellulose filter arrayed with antibodies against 400 signal transduction proteins was blotted with lysates of RPMI 8226 cells transfected with GFP-tagged PAK4. The multiprotein complexes were detected by immunoblotting with a horseradish peroxidase–conjugated anti-GFP antibody and visualized by chemiluminescence. (F) Nitrocellulose filters immobilized with 24 antibodies were incubated with whole cell lysates from cells untreated or treated with KPT-9274. Data are presented as signal intensity. TR, treated.
Using a high-throughput array containing 400 antibodies against signal transduction proteins, we have identified FGFR3 as a binding partner of PAK4 in myeloma cells (Figure 5E). Based on this observation, we next investigated whether KPT-9274 treatment could disrupt this interaction and therefore specifically sensitize FGFR3-addicted cells. Protein binding analysis revealed disruption of PAK4 interaction with FGFR3 after KPT-9274 treatment (Figure 5F), suggesting that targeting PAK4 may have a significant impact especially in FGFR3-driven myeloma.
Finally, treatment with KPT-9274 triggered a dose- and time-dependent induction of apoptosis in myeloma cells cultured in the presence or absence of BMSC, as evaluated by flow cytometric analysis following Annexin V and propidium iodide (PI) staining (Figure 6A and data not shown). Both intrinsic and extrinsic apoptotic pathways through caspases -3/7, -8, and −9 activation and poly-ADP ribose polymerase cleavage were induced in MM cells by KPT-9274, as assessed by WB analysis (Figure 6B) and luminescence assays (Figure 6C). Caspases activation by KPT-9274 treatment was also confirmed in primary CD138+ patient MM cells (Figure 6D). Exposure to the pan-caspase inhibitor zVAD significantly decreased the induction of apoptosis by KPT-9274, suggesting a caspase-dependent apoptotic process (Figure 6E).
KPT-9274 triggers apoptotic cell death in myeloma cells. (A) OPM2 cells were cultured in the absence or presence of KPT-9274, and apoptotic cell death was assessed by flow cytometric analysis following AnnexinV and PI staining. The percent of AnnexinV+/PI− (early apoptosis) and AnnexinV+/PI+ (late apoptosis) cells are shown in the graphs. (B) Whole cell lysate from OPM2 cells treated with several concentrations of KPT-9274 for 48 hours was subjected to WB analysis and probed with antibodies against caspases-3, -8, -9, poly-ADP ribose polymerase, with β-actin as loading control. (C-D) Indicated caspase activities were evaluated in OPM2 cells (C) and CD138+ cells from 4 MM patients (D) after KPT-9274 treatment (0.5 µM) for 48 and/or 72 hours using luminescence assay. (E) OPM2 cells were cultured in the presence or absence of zVAD-fmk (100 µM) with or without KPT-9274 for 48 hours. Apoptosis was evaluated by flow cytometric analysis following Annexin V and PI staining. The percent of AnnexinV+ cells is shown in the graph.
KPT-9274 triggers apoptotic cell death in myeloma cells. (A) OPM2 cells were cultured in the absence or presence of KPT-9274, and apoptotic cell death was assessed by flow cytometric analysis following AnnexinV and PI staining. The percent of AnnexinV+/PI− (early apoptosis) and AnnexinV+/PI+ (late apoptosis) cells are shown in the graphs. (B) Whole cell lysate from OPM2 cells treated with several concentrations of KPT-9274 for 48 hours was subjected to WB analysis and probed with antibodies against caspases-3, -8, -9, poly-ADP ribose polymerase, with β-actin as loading control. (C-D) Indicated caspase activities were evaluated in OPM2 cells (C) and CD138+ cells from 4 MM patients (D) after KPT-9274 treatment (0.5 µM) for 48 and/or 72 hours using luminescence assay. (E) OPM2 cells were cultured in the presence or absence of zVAD-fmk (100 µM) with or without KPT-9274 for 48 hours. Apoptosis was evaluated by flow cytometric analysis following Annexin V and PI staining. The percent of AnnexinV+ cells is shown in the graph.
KPT-9274–induced MM cell death is mediated by MEK/ERK pathway deregulation
We next evaluated and observed a significant impact of KPT-9274 on activation of MEK/ERK, AKT, and NF-κB signaling pathways (Figure 7A). In order to establish a predominant pathway responsible for the antimyeloma activity of PAK4 inhibitor, we performed NF-қB and ERK promoter activity assay in both resistant and sensitive cell lines in the presence and absence of KPT-9274. We observed inhibition of NF-қB activity in all MM cell lines tested, including the resistant U266 cell line, upon KPT-9274 treatment (Figure 7B). Strikingly, ERK activity was significantly inhibited in the sensitive MM cell lines after treatment with KPT-9274, but not in the resistant U266 cells (Figure 7C).
MEK/ERK pathway deregulation mediates KPT-9274–induced MM cell death. (A) Whole cell lysates from OPM2 cells treated with several concentrations of KPT-9274 for 48 hours were subjected to WB analysis and probed with indicated antibodies. (B) MM1S, KMS11, OPM2, and U266 cells were electroporated with control or NF-κB luciferase reporter plasmid and pRL-TK to normalize for different transfection efficiencies; following electroporation, cells were treated with vehicle or 2 doses of KPT-9274. Forty-eight hours later, luminescence was measured using the Dual Luciferase assay kit and the Glo-Max microplate luminometer. Results are expressed as percentage of Firefly/Renilla ratio of control-transfected cells. (C) H929, KMS11, and U266 cells were electroporated with control or serum response element (SRE) reporter vector. Dual Luciferase assay was performed after 48 hours of incubation with or without KPT-9274. Results are expressed as percentage of Firefly/Renilla ratio of control-transfected cells. (D) Relative mRNA expression of differentially expressed genes after KPT-9274 treatment in OPM2 and KMS11 cell lines compared with untreated cells. (E) Nuclear extracts from KPT-9274–treated OPM2 cells were analyzed for transcription factor activation using a transcription factor profiling array. Relative fold changes from control are plotted. (F) OPM2 cells were treated with different concentrations of KPT-9274 in combination with either U0126 (10 µM) or LY29004 (10 µM) or Rapamycin (10 µM) for 48 hours. Cell viability was assessed by CTG uptake and presented as percent compared with control cells.
MEK/ERK pathway deregulation mediates KPT-9274–induced MM cell death. (A) Whole cell lysates from OPM2 cells treated with several concentrations of KPT-9274 for 48 hours were subjected to WB analysis and probed with indicated antibodies. (B) MM1S, KMS11, OPM2, and U266 cells were electroporated with control or NF-κB luciferase reporter plasmid and pRL-TK to normalize for different transfection efficiencies; following electroporation, cells were treated with vehicle or 2 doses of KPT-9274. Forty-eight hours later, luminescence was measured using the Dual Luciferase assay kit and the Glo-Max microplate luminometer. Results are expressed as percentage of Firefly/Renilla ratio of control-transfected cells. (C) H929, KMS11, and U266 cells were electroporated with control or serum response element (SRE) reporter vector. Dual Luciferase assay was performed after 48 hours of incubation with or without KPT-9274. Results are expressed as percentage of Firefly/Renilla ratio of control-transfected cells. (D) Relative mRNA expression of differentially expressed genes after KPT-9274 treatment in OPM2 and KMS11 cell lines compared with untreated cells. (E) Nuclear extracts from KPT-9274–treated OPM2 cells were analyzed for transcription factor activation using a transcription factor profiling array. Relative fold changes from control are plotted. (F) OPM2 cells were treated with different concentrations of KPT-9274 in combination with either U0126 (10 µM) or LY29004 (10 µM) or Rapamycin (10 µM) for 48 hours. Cell viability was assessed by CTG uptake and presented as percent compared with control cells.
Gene expression profiling in the OPM2 and KMS11 cell lines after KPT-9274 treatment confirmed a significant decrease of ERK target genes, as well as antiapoptotic and FGFR3-related genes (MCL1 and DUSP6 among the others) compared with untreated cells (Figure 7D) (Gene Expression Omnibus accession number GSE93745). These observations were also confirmed by qPCR and WB analysis in additional myeloma cell lines (data not shown). Interestingly, ingenuity pathway analysis confirmed the presence of 2 major gene network hubs: MEK (activation z-score −2.8) and MYC (activation z-score −2.2).
Intriguingly, we have evidence that PAMs also inhibit the enzymatic activity of nicotinamide phosphoribosyltransferase (NAMPT; supplemental Figure 6A-B). We found that NAMPT forms a complex with the conserved PAK binding domain independently of Cdc42 activation, and treatment with KPT-9274 disrupted this interaction ( supplemental Figure 6C-D). Cea et al have shown that nicotinamide adenine dinucleotide (NAD) depletion decreased myeloma cell proliferation through ERK inhibition.25 Our observations suggest that PAK4 and the NAMPT pathway may cooperate to regulate ERK signaling in myeloma, which can be efficiently targeted by KPT-9274.
ERKs can directly phosphorylate and therefore activate a set of transcription factors to regulate a diverse range of cellular processes.26 By using a high throughput transcription factor DNA binding assay, we observed significant decreased binding to the DNA domains of established nuclear targets of activated ERK, including AP1, Ets, EGR1, and CREB, in myeloma cells upon treatment with KPT-9274 (Figure 7E). These results establish PAK4 as an oncogenic activator of MEK that induces phosphorylation and therefore induction of ERK1/2 signaling pathway. This intersection between PAK4 and the MEK/ERK signaling may ultimately impact expression and/or activity of downstream targets endowed with oncogenic potential in myeloma, such as MYC and Cyclin D2. Importantly, AKT and mTOR inhibitors were synergistic with KPT-9274, while the combination with the MEK inhibitor U0126 was at best ineffective or nearly antagonistic (Figure 7F; supplemental Figure 4G).
Altogether, this set of data establishes PAK4 as an oncogenic activator of MEK/ERK signaling pathway in myeloma cells that can be therapeutically targeted with KPT-9274.
Discussion
STKs play an important role in cellular homeostasis and signaling through their ability to phosphorylate transcription factors, cell cycle regulators, and a vast array of cytoplasmic and nuclear effectors.27 A number of STKs have been implicated in human cancers, including myeloma.28 The PAK family of STKs has been associated with malignant transformation. Although PAK1 and PAK2 have been shown to be relevant in myeloma cell migration,29 the role of PAK4 in MM remains unknown. The PAK4 gene lies within chromosome region 19q13.2 commonly amplified in 30% of MM patients.30 We confirmed a correlation between copy number amplification and increased expression of PAK4 in 2 different myeloma patient datasets (data not shown). Moreover, high expression of total and phosphorylated PAK4 was also observed in the premalignant monoclonal gammopathy of undetermined significance and in the SMM stages, suggesting that its overexpression may be an early event in the pathogenesis of myeloma.
PAK4 is involved in a wide range of biological activities, which include protecting cells against apoptosis, activating cell survival pathways under stress, inhibiting cell adhesion, and promoting cell migration.6,7,17,19,31-33 In this study, we show that myeloma cells with high PAK4 expression display sensitivity to conditional PAK4 knockdown, implying that an oncogene-addicted state exists in such cells. Moreover, we show that PAK4 promotes myeloma cell proliferation in vitro as well as in vivo in humanized murine models of myeloma through activation of antiapoptotic and survival signaling pathways, including the MEK/ERK pathway. This observation is further highlighted by sensitivity to a novel small molecule inhibitor of PAK4, KPT-9274, that has recently entered a phase 1 clinical trial in patients with solid malignancies or non-Hodgkin lymphoma (NCT02702492).
KPT-9274 effectively inhibits MM cell growth and survival, with no significant effect on normal PBMCs or bone marrow mononuclear cells, suggesting a potentially favorable therapeutic index. Inhibition of ERK by KPT-9274 also correlates with decreased DNA binding activity of ERK-dependent transcription factors AP1, ETS, CREB, and EGR, and decreased expression of ERK target genes such as CCND2, CCR1, and MYC. The observed lack of synergistic/additive effect of KPT-9274 in combination with MEK inhibitor U0126 (supplemental Figure 4G) further confirms ERK as one of the major pathway responsible for its activity in myeloma.
Intriguingly, we have evidence that PAMs also inhibit the enzymatic activity of NAMPT, by disrupting the interaction between the conserved PAK binding domain and NAMPT. We have previously shown that MM cells are remarkably dependent on NAMPT activity25 and observed a significant cytotoxic activity against tumor cells via inhibition of ERK and mTOR pathways by FK866 (APO866), identifying NAD depletion as a promising antimyeloma therapy. Our observations suggest that PAK4 and the NAMPT (NAD) salvage pathway may cooperate to regulate ERK signaling. Therefore, this dual activity of KPT-9274 against PAK4 and NAMPT may provide a higher level of ERK pathway inhibition compared with drugs targeting individual pathways alone.
Finally, we have identified FGFR3, a commonly disrupted tyrosine kinase receptor,34,35 and Grb2, an adaptor protein involved in RAS activation,37 as novel PAK4 binding partners in myeloma and report disruption of this binding by KPT-9274. As a result, t(4;14)-positive MM cells expressing FGFR3 show greater sensitivity to PAK4 inhibition compared with MM cell lines without FGFR3 aberrations (eg, U266 and ANBL6). These observations are in line with the observed effect of KPT-9274 on MEK-ERK signaling, a pathway also induced by activating FGFR3 mutation.36 However, FGFR3 downstream signaling pathways inducing oncogenesis and cancer cell growth are not yet fully delineated. A detailed investigation of cellular and molecular impact of FGFR3-PAK4 interaction may therefore provide new insights on FGFR3-mediated oncogenic signaling pathways and its therapeutic potential. Our findings indicate that FGFR3 may serve as a predictive marker of KPT-9274 sensitivity in MM patients, with the potential for broader application to other malignancies associated with dysregulation of FGFR3, including human bladder and cervical carcinomas.38
In conclusion, our study sheds light on the oncogenic role of the STK PAK4 as survival and antiapoptotic factor in myeloma and shows that its inhibition with a new allosteric modulator, KPT-9274, represents a potential novel therapeutic intervention in MM, especially in high-risk t(4:14)-positive patients.
The data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE93745).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the Ron and Anita Wornick Fund, National Institutes of Health National Cancer Institute grants PO1-155258 and P50-100707 (N.C.M. and K.C.A.), Department of Veterans Affairs Merit Review Award 1 I01BX001584-01 (N.C.M.), and BA15/00035 and Cancer Research Innovation Spain grants (J.M.-L.).
Authorship
Contribution: M.F. and N.C.M. designed the study and wrote the manuscript; M.F., J.M.-L., W.S., S.O., and R.L.B. performed the experiments; M.F. and J.M.-L. analyzed the data; N.A., R.S., A.R., and M.C. provided vital reagents and inputs to the study; Y.X. and A.G. performed protein binding assay; M.L. performed IHC staining; M.K.S. analyzed gene expression data; E.B. and S.S. designed KPT-9274 and its analogs; C.A. performed the PAK-NAMPT pull-down experiments; Y.L. and B.K. planned and designed the stable isotope labeling of amino acids in cells experiments; S.L., S.-L.W., and M.S. performed mass spectrometry runs and data analysis; and W.S., R.P., and K.C.A. provided critical evaluation of experimental data and manuscript.
Conflict-of-interest disclosure: W.S., E.B., S.S., C.A., and Y.L. are employees of Karyopharm Therapeutics Inc. N.C.M. serves on advisory boards to Millennium, Celgene, and Novartis. K.C.A. serves on advisory boards to Celgene, Millennium, BMS, and Gilead and holds equity ownership in Oncopep and Acetylon. The remaining authors declare no competing financial interests.
Correspondence: Nikhil C. Munshi, Dana-Farber Cancer Institute, 440 Brookline Ave, M230, Boston, MA 02115; e-mail: nikhil_munshi@dfci.harvard.edu.
References
Author notes
M.F. and J.M.-L. contributed equally to this study.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal