Key Points
Most cases of ibrutinib-resistant CLL were due to mutations in BTK and/or PLCG2 and often composed of multiple independent subclones.
High sensitivity testing identified resistance mutations up to 15 months before manifestation of clinical progression.
Abstract
Disease progression in patients with chronic lymphocytic leukemia (CLL) treated with ibrutinib has been attributed to histologic transformation or acquired mutations in BTK and PLCG2. The rate of resistance and clonal composition of PD are incompletely characterized. We report on CLL patients treated with single-agent ibrutinib on an investigator-initiated phase 2 trial. With median follow-up of 34 months, 15 of 84 evaluable patients (17.9%) progressed. Relapsed/refractory disease at study entry, TP53 aberration, advanced Rai stage, and high β-2 microglobulin were independently associated with inferior progression-free survival (P < .05 for all tests). Histologic transformation occurred in 5 patients (6.0%) and was limited to the first 15 months on ibrutinib. In contrast, progression due to CLL in 10 patients (11.9%) occurred later, diagnosed at a median 38 months on study. At progression, mutations in BTK (Cys481) and/or PLCG2 (within the autoinhibitory domain) were found in 9 patients (10.7%), in 8 of 10 patients with progressive CLL, and in 1 patient with prolymphocytic transformation. Applying high-sensitivity testing (detection limit ∼1 in 1000 cells) to stored samples, we detected mutations up to 15 months before manifestation of clinical progression (range, 2.9-15.4 months). In 5 patients (6.0%), multiple subclones carrying different mutations arose independently, leading to subclonal heterogeneity of resistant disease. For a seamless transition to alternative targeted agents, patients progressing with CLL were continued on ibrutinib for up to 3 months, with 19.8 months median survival from the time of progression. This trial was registered at www.clinicaltrials.gov as #NCT01500733.
Introduction
Chronic lymphocytic leukemia (CLL) is characterized by the clonal expansion of autoreactive B cells whose proliferation and survival are dependent on the tissue microenvironment and B-cell receptor (BCR) signaling.1-4 Ibrutinib covalently binds to Bruton tyrosine kinase (BTK) and inhibits BCR and downstream NF-κB signaling in CLL cells in vivo, leading to reduced tumor proliferation and increased cell death.5,6 In 2 randomized phase 3 trials, ibrutinib improved survival of CLL patients with relapsed/refractory and previously untreated diseases when compared with ofatumumab and chlorambucil, respectively.7,8 Response rates to single-agent ibrutinib are high and independent of prior treatment status reaching 63% to 88% in relapsed/refractory CLL8,9 and 71% to 86% in previously untreated patients.7,10 In CLL with del(17p), ibrutinib achieves superior results compared with historic data with chemotherapy in both frontline and relapsed/refractory settings.11 For most published studies, follow-up is limited to 2 years or less. The initial phase 1b/2 study, recently updated with up to 3 years of follow-up, reported durable response in most patients.12 However, patients with relapsed/refractory disease and del(17p) were more likely to progress and had a median progression-free survival (PFS) of 28.1 months. We recently reported PFS of 82% at 24 months for patients with TP53 aberrations treated with ibrutinib in first-line or beyond.11 In comparison, median PFS for fludarabine, cyclophosphamide plus rituximab (FCR),13 and of bendamustine plus rituximab14 has been reported as 11.3 months and 7.9 months, respectively, in first-line for CLL with del(17p). Ibrutinib gained initial regulatory approval in 2014 for relapsed/refractory CLL and CLL with del(17p) that was recently expanded to include all patients with CLL.
A possible limitation of single-agent therapy is the emergence of drug resistance. Progression on ibrutinib appears to fall into 2 distinct patterns.15 Patients with primary refractory disease or early progression often present with histologic transformation.11,16,17 In contrast, delayed progression after initial response commonly appears as CLL harboring acquired mutations. In a study of 11 patients who progressed with CLL, all tested cases showed either BTK Cys481 mutations, which prevent the covalent binding of ibrutinib, or PLCG2 gain-of-function mutations that activate phospholipase Cγ 2 (PLCγ2) downstream of BTK.17-19 More recently, del(8p), leading to haploinsufficiency of TRAIL-R, in conjunction with additional driver mutations in EP300, MLL2, and EIF2A has been described in patients developing ibrutinib resistance.20 Important unanswered questions about BTK and PLCG2 mutations include the prevalence of mutations over time, the identification of at-risk populations, and how the presence of mutations affects disease behavior and response to salvage treatment. Here, we report on the clinical and molecular characteristics of progressive disease developing during longitudinal follow-up of 84 CLL patients who were treated with single-agent ibrutinib over a median 3 years; 53 of whom (63.1%) had TP53 aberration.
Patients, materials, and methods
Patients and study design
Eighty-six CLL patients were treated with single-agent ibrutinib under a phase 2 investigator-initiated trial (clinicaltrials.gov #NCT01500733). Eligible patients had either TP53 aberration (del(17p) or TP53 mutation) or were ≥65 years old. Two patients were removed from the study for enrollment deviations. Interphase fluorescence in situ hybridization (FISH) was done prior to ibrutinib therapy in all cases; G-banded karyotype was not routinely performed. Eligible patients had Eastern Cooperative Oncology Group performance status ≤2 and adequate organ function. All patients received ibrutinib 420 mg once daily until disease progression or intolerable side effects. The study was approved by the institutional review board at the National Heart, Lung, and Blood Institute, and conducted in accordance with the Declaration of Helsinki. All patients provided written informed consents.
Definition of treatment response and disease progression
The primary study end point was treatment response at 6 months according to 2008 revised International Workshop on Chronic Lymphocytic Leukemia (IWCLL) criteria,21 incorporating the 2012 clarifications for patients treated with kinase inhibitors as previously reported.11 Response assessments at 2, 6, and 12 months and annually thereafter included physical and laboratory examinations, computed tomography imaging, bone marrow biopsy, and flow cytometry to quantify residual disease as described.22 Progression in patients who initially responded to treatment required the absence of confounding factors, including recent interruptions of drug therapy, and was based on 2008 IWCLL criteria but using best response (“nadir”) instead of baseline as the reference (supplemental Table 1, available on the Blood Web site).23 At progression, flow cytometry, bone marrow biopsies, and/or lymph node biopsies were performed for diagnosis or exclusion of disease transformation.
Sequencing of BTK and PLCG2 genes
Genomic DNA from peripheral blood or bone marrow samples of patients who progressed was tested for mutations in BTK and PLCG2 using previously described methods.24,25 Briefly, exon 15 of BTK and exons 19, 20, and 24 of PLCG2 were amplified using custom oligonucleotides and analyzed by bidirectional Sanger sequencing. For high-sensitivity assays, wild-type blocking polymerase chain reactions (PCRs) using branched or locked nucleic acids followed by a high-sensitivity next-generation sequencing (NGS) panel of 315 genes was used (NeoGenomics). High-sensitivity NGS is able to detect a 0.1% mutant allele in the background of wild type. In patients with detectable mutations at progression, stored samples from earlier time points were also sequenced.
Droplet digital PCR
Droplet digital PCR (ddPCR) was performed on DNA isolated from stored samples after CD19+ selection, as described previously.26 Briefly, custom ddPCR assays were obtained from Bio-Rad Laboratories and analyzed using a Bio-Rad QX200 droplet reader (Hercules, CA). Each mutation detection assay was run duplexed to a matched wild-type assay (primer and probe sequences, supplemental Table 2). All experiments were performed in either triplicates or quadruplicates. Variant allele frequencies (VAFs) were calculated from the fraction of droplets containing at least 1 variant allele with a reported sensitivity of 0.01%. Analytical specificity for assays reporting VAFs <1% was tested using DNA from peripheral blood leukocytes of 12 normal donors obtained from Biochain (San Francisco, CA) and computed as the percentage of normal donor samples having VAFs equal or less than the patients’ sample. Assuming heterozygous mutations, the clonal burden was estimated from the VAF multiplied by 2 (multiplied by 1 for X-chromosomal BTK in male patients) and the percentage of CD19+ cells among lymphocytes.
Serum levels of CCL3 and CCL4
Serum was stored at −80°C until analysis. Concentrations of CCL3 and CCL4 were measured using the HCYTMAG-60K-PX38 custom-designed assay (Millipore, Billerica, MA) with the Luminex 200 instrument (Austin, TX) according to the manufacturer’s instructions.
Statistical analysis
PFS was defined as the time from the start of ibrutinib until progression or death. Probability of PFS was estimated by the Kaplan-Meier method, and patients alive and progression-free were censored at the last follow-up.27 The log-rank test was used to compare PFS probabilities between subgroups. Regression models were used to identify factors associated with progression. Time to progression (TTP) was defined from the start of ibrutinib until disease progression, and the cumulative incidence of progression was estimated with deaths without progression considered as competing risk events.28 All tests were 2-sided and P values <.05 were considered statistically significant. Statistical analyses were performed using R statistical software version 3.2.3 (R Foundation for Statistical Computing).
Results
Incidence and biology of progressive disease
With a median follow-up of 34.3 months (0.1-49.7), 4 of 84 patients (4.8%) died on study: 3 due to infections and 1 patient with sudden death of unknown cause. Fifteen patients (17.9%) progressed, using best response as the new baseline. Seven patients (8.3%) discontinued treatment due to adverse events or patient choice and 58 (69.0%) continue on ibrutinib.
Of the 15 patients with progression, 2 had primary refractory disease and progressed within the first 2 cycles of therapy, 3 had stable diseases for at least 6 months, and 10 patients achieved objective responses before progression. At progression, 7 patients (46.6%) presented with ≥50% increase in sum of the product of the diameters of representative lymph nodes with or without increase in absolute lymphocyte count (ALC) (supplemental Table 1). In 3 additional patients (20.2%), progression was defined by ≥50% increase in ALC confirmed in 2 consecutive assessments and with an absolute B-cell count >5000/μL. The remaining 5 patients (33.3%) developed a new lymph node or histologic transformation at progression.
Table 1 summarizes clinical characteristics of the patients who progressed (PD group) and those who did not (non-PD group). A higher percentage of patients in the PD group than the non-PD group had del(17p), high β-2 microglobulin (B2M; >4 mg/L), and relapsed or refractory disease (supplemental Table 3).
Patient characteristics
| . | PD, n = 15 . | Non-PD, n = 69 . | P* . |
|---|---|---|---|
| Age, median (range), y | 65 (56-79) | 68 (33-85) | .39 |
| Sex, n (%) | .99 | ||
| Male | 9 (60.0) | 40 (58.0) | |
| Female | 6 (40.0) | 29 (42.0) | |
| Rai stage, n (%) | .36 | ||
| Rai I/II | 3 (20.0) | 24 (34.8) | |
| Rai III/IV | 12 (80.0) | 45 (65.2) | |
| FISH, n (%)† | .04‡ | ||
| Deletion 13q | 0 (0.0) | 10 (14.5) | |
| Normal | 0 (0.0) | 1 (1.4) | |
| Trisomy 12 | 2 (13.3) | 7 (10.1) | |
| Deletion 11q | 0 (0.0) | 11 (15.9) | |
| Deletion 17p | 13 (86.7) | 40 (58.0) | |
| IGHV, n (%) | .24 | ||
| Mutated | 3 (20.0) | 26 (37.7) | |
| Unmutated | 12 (80.0) | 43 (62.3) | |
| B2M | |||
| Median (range)§ | 5.7 (1.8-10.3) | 3.6 (1.7-12.9) | .005 |
| >4 mg/L, n (%) | 13 (86.7) | 27 (39.1) | .001 |
| >3.5 mg/L, n (%) | 13 (86.7) | 35 (50.7) | .02 |
| Prior treatment, n (%) | .003 | ||
| Treatment-naive | 4 (26.7) | 48 (69.6) | |
| Relapsed/refractory | 11 (73.3) | 21 (30.4) |
| . | PD, n = 15 . | Non-PD, n = 69 . | P* . |
|---|---|---|---|
| Age, median (range), y | 65 (56-79) | 68 (33-85) | .39 |
| Sex, n (%) | .99 | ||
| Male | 9 (60.0) | 40 (58.0) | |
| Female | 6 (40.0) | 29 (42.0) | |
| Rai stage, n (%) | .36 | ||
| Rai I/II | 3 (20.0) | 24 (34.8) | |
| Rai III/IV | 12 (80.0) | 45 (65.2) | |
| FISH, n (%)† | .04‡ | ||
| Deletion 13q | 0 (0.0) | 10 (14.5) | |
| Normal | 0 (0.0) | 1 (1.4) | |
| Trisomy 12 | 2 (13.3) | 7 (10.1) | |
| Deletion 11q | 0 (0.0) | 11 (15.9) | |
| Deletion 17p | 13 (86.7) | 40 (58.0) | |
| IGHV, n (%) | .24 | ||
| Mutated | 3 (20.0) | 26 (37.7) | |
| Unmutated | 12 (80.0) | 43 (62.3) | |
| B2M | |||
| Median (range)§ | 5.7 (1.8-10.3) | 3.6 (1.7-12.9) | .005 |
| >4 mg/L, n (%) | 13 (86.7) | 27 (39.1) | .001 |
| >3.5 mg/L, n (%) | 13 (86.7) | 35 (50.7) | .02 |
| Prior treatment, n (%) | .003 | ||
| Treatment-naive | 4 (26.7) | 48 (69.6) | |
| Relapsed/refractory | 11 (73.3) | 21 (30.4) |
The Fisher exact test and the Wilcoxon rank-sum test were used to compare categorical variables and continuous variables between patients who progressed (PD) and those who did not (non-PD).
Hierarchical FISH categories.
The Fisher exact test in subgroups divided by the presence or the absence of deletion 17p.
The median B2M for all patients was 4 mg/L.
Of 15 patients who progressed, 5 had disease transformation and 10 progressed with CLL with or without a prolymphocytic component (Table 2). Four patients were considered to have Richter transformation; biopsy confirmed diffuse large B-cell lymphoma in 2 patients, and in 1 patient each, Epstein-Barr virus–positive (EBV+) Hodgkin lymphoma and accelerated CLL with interspersed Hodgkin-like cells. One patient progressed within 2 weeks of starting ibrutinib with a clinical presentation consistent with Richter transformation; however, tissue biopsy was not obtained. The median TTP was 27.9 months (0.4-47.4). The last case of transformation was diagnosed at 15.2 months. All subsequent progression events were due to CLL.
Histopathology and TTP
| Year . | CLL without transformation (mo) . | Transformed . | Total . |
|---|---|---|---|
| <1 | PD3 (5.6) | PD1 (0.4 mo, suspected Richter transformation) | 4 |
| PD2 (1.1 mo, EBV+ HL) | |||
| PD4 (8.4 mo, DLBCL) | |||
| 1 to <2 | PD7 (23.8) | PD5 (14.7 mo, HL/CLL) | 3 |
| PD6 (15.2 mo, DLBCL) | |||
| 2 to <3 | PD8 (27.9) | 2 | |
| PD9 (33.9) | |||
| ≥3 | PD10 (36.9) | 6 | |
| PD11 (38.4) | |||
| PD12 (38.8) | |||
| PD13 (38.9) | |||
| PD14 (42.8) | |||
| PD15 (47.4) | |||
| Total | 10 | 5 | 15 |
| Year . | CLL without transformation (mo) . | Transformed . | Total . |
|---|---|---|---|
| <1 | PD3 (5.6) | PD1 (0.4 mo, suspected Richter transformation) | 4 |
| PD2 (1.1 mo, EBV+ HL) | |||
| PD4 (8.4 mo, DLBCL) | |||
| 1 to <2 | PD7 (23.8) | PD5 (14.7 mo, HL/CLL) | 3 |
| PD6 (15.2 mo, DLBCL) | |||
| 2 to <3 | PD8 (27.9) | 2 | |
| PD9 (33.9) | |||
| ≥3 | PD10 (36.9) | 6 | |
| PD11 (38.4) | |||
| PD12 (38.8) | |||
| PD13 (38.9) | |||
| PD14 (42.8) | |||
| PD15 (47.4) | |||
| Total | 10 | 5 | 15 |
Patient identifier (TTP, histology if transformed).
DLBCL, diffuse large B-cell lymphoma; EBV+ HL, EBV+ Hodgkin lymphoma; HL/CLL, Hodgkin-like cells mixed with CLL.
Progression-free survival
The estimated 36-month PFS was 83.6% (95% confidence interval [CI], 75.7%-92.3%; Figure 1A). PFS was significantly inferior in subgroups with TP53 aberration (P = .026), relapsed or refractory disease status (P = .025), advanced Rai stage (III/IV) (P = .047), and high B2M (>4 mg/L) (P = .013), compared with their respective counterparts (Figure 1B-E). B2M of 4 mg/L was the median value of the study population, and was thus selected as a cutoff to dichotomize this variable. Patients having immunoglobulin gene heavy-chain variable region (IGHV)-unmutated CLL tended to have shorter PFS than those with IGHV-mutated disease; however, this was not statistically significant (P = .12; Figure 1F).
PFS. Kaplan-Meier estimates of PFS of (A) all patients and subgroups stratified by: (B) TP53 aberration (absent vs present), (C) prior treatment (RR CLL vs TN), (D) Rai stage (I/II vs III/IV), (E) B2M (≤4 mg/L vs >4 mg/L), and (F) IGHV mutational status (mutated IGHV [M] vs unmutated IGHV [U]). Among patients with TP53 aberration (n = 53), subgroups are further divided by: (G) prior treatment (RR vs TN), (H) Rai stage (I/II vs III/IV), and (I) B2M (≤4 mg/L vs >4 mg/L).
PFS. Kaplan-Meier estimates of PFS of (A) all patients and subgroups stratified by: (B) TP53 aberration (absent vs present), (C) prior treatment (RR CLL vs TN), (D) Rai stage (I/II vs III/IV), (E) B2M (≤4 mg/L vs >4 mg/L), and (F) IGHV mutational status (mutated IGHV [M] vs unmutated IGHV [U]). Among patients with TP53 aberration (n = 53), subgroups are further divided by: (G) prior treatment (RR vs TN), (H) Rai stage (I/II vs III/IV), and (I) B2M (≤4 mg/L vs >4 mg/L).
Among the subgroup of 53 patients with TP53 aberration, relapsed/refractory disease (hazard ratio [HR], 2.67), advanced Rai stage (HR, 2.99), and high B2M (HR, 3.41) remained to show a trend toward inferior outcome (Figures 1G-I). The cumulative incidence of progression was significantly higher in patients with relapsed/refractory CLL (RR) than previously untreated CLL (treatment-naive [TN]) (47% in RR group vs 15% in TN group at 48 months). The TTP was significantly shorter in the RR group compared with the TN group (P = .027) (supplemental Figure 1).
We performed multivariate Cox regression analyses of PFS and 5 risk factors such as age, IGHV mutation status, TP53 aberration, Rai stage, and prior treatment status. TP53 aberration, advanced Rai stage (III/IV), and relapsed/refractory disease were independently associated with higher risk of progression after adjusting for age and IGHV status at enrollment.
Resistance mutations at disease progression
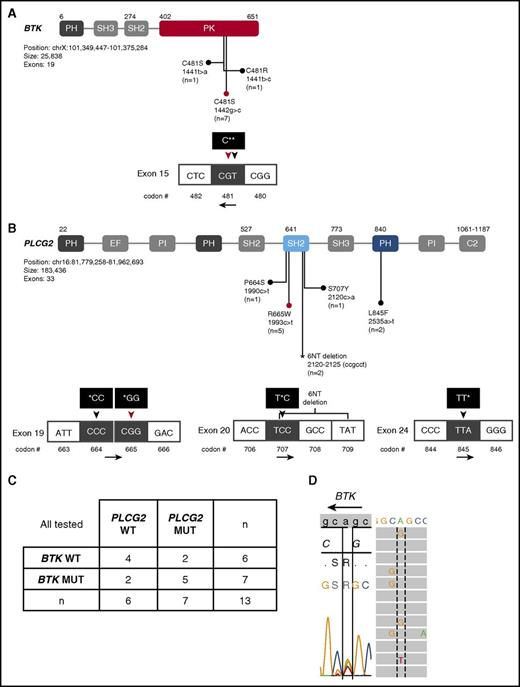
Mutations previously identified in CLL patients who progressed on ibrutinib affect exon 15 of BTK (C481S) and exons 19 (R665W), 20 (S707F, P or Y), and 24 (L845F) of PLCG2.17,18,29 We sequenced these candidate regions in 13 patients with available peripheral blood or bone marrow samples at progression. One of 3 cases with histologic transformation had a mutation at PLCG2 (R665W), whereas 8 of 10 cases with progressive CLL had BTK and/or PLCG2 mutations (Table 3). Seven distinct nonsynonymous mutations were identified (Figure 2A-B); 3 different nonsynonymous mutations in BTK exon 15 (c.1441T>A [C481S], c.1441T>C [C481R], c.1442G>C [C481S]) and 4 types of nonsynonymous mutations in PLCG2 exon 19 (c.1990C>T [P664S], c.1993C>T [R665W]), exon 20 (c.2120C>A [S707Y]), and exon 24 (c.2535A>T [L845F]). Additionally, 2 patients had a 6-nucleotide deletion in exon 20 of PLCG2 (c.2120-2125del), leading to the deletion of S707 and A708. This deletion and the PLCG2 c.1990C>T (P664S) mutation have not been previously reported in CLL; however, both events affected the autoinhibitory Src homology 2 (SH2) domain of PLCγ2, the recurrent target of previously identified mutations. Two patients who progressed with CLL after 3 years on ibrutinib did not have any detectable mutation in the candidate regions of BTK or PLCG2.
Mutations of BTK and PLCG2 genes
| Subject . | Gene . | Exon . | AA change . | WT codon . | MT codon* . | Previously reported . | VAF, %† . |
|---|---|---|---|---|---|---|---|
| PD1‡ | Not tested | — | — | — | — | — | — |
| PD2‡ | Not tested | — | — | — | — | — | — |
| PD3 | BTK | 15 | C481S | TGC | TCC | Yes | § |
| PLCG2 | 20 | 6NT deletion | — | — | No | § | |
| PD4‡ | None detected | — | — | — | — | — | — |
| PD5 | None detected | — | — | — | — | — | — |
| PD6‡ | PLCG2 | 19 | R665W | CGG | TGG | Yes | 0.11 |
| PD7 | PLCG2 | 19 | R665W | CGG | TGG | Yes | ND |
| PLCG2 | 19 | P664S | CCC | TCC | No | ND | |
| PD8‡ | BTK | 15 | C481S | TGC | TCC | Yes | 78.2 |
| PLCG2 | 19 | R665W | CGG | TGG | Yes | 0.26 | |
| PLCG2 | 20 | S707Y | TCC | TAC | Yes | 0.17 | |
| PLCG2 | 24 | L845F | TTA | TTT | Yes | 4.7 | |
| PD9‡ | BTK | 15 | C481S | TGC | TCC | Yes | 8.8 |
| BTK | 15 | C481S | TGC | AGC | Yes | 7.2 | |
| BTK | 15 | C481R | TGC | CGC | Yes | 15.8 | |
| PLCG2 | 19 | R665W | CGG | TGG | Yes | 7.3 | |
| PLCG2 | 24 | L845F | TTA | TTT | Yes | 18.3 | |
| PD10 | BTK | 15 | C481S | TGC | TCC | Yes | 1.6 |
| PLCG2 | 19 | R665W | CGG | TGG | Yes | 0.1 | |
| PD11‡ | BTK | 15 | C481S | TGC | TCC | Yes | 57.3 |
| PD12 | None detected | — | — | — | — | — | — |
| PD13‡ | BTK | 15 | C481S | TGC | TCC | Yes | 32.6 |
| PLCG2 | 20 | 6NT deletion | — | — | No | ND | |
| PD14 | None detected | — | — | — | — | — | — |
| PD15‡ | BTK | 15 | C481S | TGC | TCC | Yes | 2.2 |
| Subject . | Gene . | Exon . | AA change . | WT codon . | MT codon* . | Previously reported . | VAF, %† . |
|---|---|---|---|---|---|---|---|
| PD1‡ | Not tested | — | — | — | — | — | — |
| PD2‡ | Not tested | — | — | — | — | — | — |
| PD3 | BTK | 15 | C481S | TGC | TCC | Yes | § |
| PLCG2 | 20 | 6NT deletion | — | — | No | § | |
| PD4‡ | None detected | — | — | — | — | — | — |
| PD5 | None detected | — | — | — | — | — | — |
| PD6‡ | PLCG2 | 19 | R665W | CGG | TGG | Yes | 0.11 |
| PD7 | PLCG2 | 19 | R665W | CGG | TGG | Yes | ND |
| PLCG2 | 19 | P664S | CCC | TCC | No | ND | |
| PD8‡ | BTK | 15 | C481S | TGC | TCC | Yes | 78.2 |
| PLCG2 | 19 | R665W | CGG | TGG | Yes | 0.26 | |
| PLCG2 | 20 | S707Y | TCC | TAC | Yes | 0.17 | |
| PLCG2 | 24 | L845F | TTA | TTT | Yes | 4.7 | |
| PD9‡ | BTK | 15 | C481S | TGC | TCC | Yes | 8.8 |
| BTK | 15 | C481S | TGC | AGC | Yes | 7.2 | |
| BTK | 15 | C481R | TGC | CGC | Yes | 15.8 | |
| PLCG2 | 19 | R665W | CGG | TGG | Yes | 7.3 | |
| PLCG2 | 24 | L845F | TTA | TTT | Yes | 18.3 | |
| PD10 | BTK | 15 | C481S | TGC | TCC | Yes | 1.6 |
| PLCG2 | 19 | R665W | CGG | TGG | Yes | 0.1 | |
| PD11‡ | BTK | 15 | C481S | TGC | TCC | Yes | 57.3 |
| PD12 | None detected | — | — | — | — | — | — |
| PD13‡ | BTK | 15 | C481S | TGC | TCC | Yes | 32.6 |
| PLCG2 | 20 | 6NT deletion | — | — | No | ND | |
| PD14 | None detected | — | — | — | — | — | — |
| PD15‡ | BTK | 15 | C481S | TGC | TCC | Yes | 2.2 |
—, not applicable; AA, amino acid; MT, mutant; ND, not detected; NT, nucleotide; WT, wild type.
Mutations were detected by Sanger sequencing or NGS as described in “Patients, materials, and methods.”
Estimated VAFs were determined by ddPCR. Specificities for VAFs <1% were tested in normal donor samples and were: 83.3% (PD6, PD10) for PLCG2 R665W and 100% (PD8) for PLCG2 S707Y.
Male patients with 1 X-chromosomal BTK allele.
Test not performed due to insufficient samples or probes.
BTK and PLCG2 mutations at disease progression. Schematic representation of functional domains of (A) BTK and (B) PLCG2 with amino acid substitutions due to nonsynonymous mutations or deletion indicated. (A) BTK gene domains and nucleotide changes. Mutations in exon 15 of BTK affect C481 in the protein tyrosine kinase (PK) domain. BTK c.1442G>C (red circle and red triangle) and c.1441T>A mutations lead to C481S. BTK c.1441T>C mutation leads to C481R substitution. (B) PLCG2 gene domains and nucleotide changes. Mutations in PLCG2 exons 19 and 20 affect the N-terminal SH2 domain and mutations in exon 24 the pleckstrin homology (PH) domain. The P664S mutation and the 6-nucleotide deletion in PLCG2 exon 20 (*), have not been previously described in CLL. (C) Number of patients with BTK and/or PLCG2 mutations at progression. (D) Sanger sequencing and NGS of patient PD9 reveals 3 different types of nucleotide changes in BTK exon 15. Shown is the sense DNA strand. BTK is encoded on the antisense strand; the black arrow indicates the read direction. Dotted and solid lines are aligned at BTK nucleotide position 1441. Left panel, The result of Sanger sequencing showing c.1442G>T (C481S), c.1441T>C (C481R), and c.1441T>A (C481S). Right panel, The result of NGS showing c.1441T>C (C481R) and c.1441T>A (C481S). EF, EF-hand domain; MUT, mutated; PI, phosphatidylinositol-specific phospholipase C X domain; WT, wild-type.
BTK and PLCG2 mutations at disease progression. Schematic representation of functional domains of (A) BTK and (B) PLCG2 with amino acid substitutions due to nonsynonymous mutations or deletion indicated. (A) BTK gene domains and nucleotide changes. Mutations in exon 15 of BTK affect C481 in the protein tyrosine kinase (PK) domain. BTK c.1442G>C (red circle and red triangle) and c.1441T>A mutations lead to C481S. BTK c.1441T>C mutation leads to C481R substitution. (B) PLCG2 gene domains and nucleotide changes. Mutations in PLCG2 exons 19 and 20 affect the N-terminal SH2 domain and mutations in exon 24 the pleckstrin homology (PH) domain. The P664S mutation and the 6-nucleotide deletion in PLCG2 exon 20 (*), have not been previously described in CLL. (C) Number of patients with BTK and/or PLCG2 mutations at progression. (D) Sanger sequencing and NGS of patient PD9 reveals 3 different types of nucleotide changes in BTK exon 15. Shown is the sense DNA strand. BTK is encoded on the antisense strand; the black arrow indicates the read direction. Dotted and solid lines are aligned at BTK nucleotide position 1441. Left panel, The result of Sanger sequencing showing c.1442G>T (C481S), c.1441T>C (C481R), and c.1441T>A (C481S). Right panel, The result of NGS showing c.1441T>C (C481R) and c.1441T>A (C481S). EF, EF-hand domain; MUT, mutated; PI, phosphatidylinositol-specific phospholipase C X domain; WT, wild-type.
In progressive CLL, we often found subclonal mutations that appeared to arise concurrently in different cells or on different alleles. Notably, in 1 patient (PD9), at least 3 separate subclones emerged that carried unique nucleotide changes in the codon for Cys481 (c.1441T>A [C481S], c.1441T>C [C481R], c.1442G>C [C481S]; Figure 2D), (Figure 2A-B). Patient PD9 is male and carries only 1 X-chromosomal BTK allele. The demonstration of 3 distinct BTK variants by NGS (Figure 2D) therefore indicates branching clonal evolution with at least 3 BTK mutant subclones evolving in parallel. This same patient also had 2 distinct PLCG2 mutations. Overall, we found concurrent BTK and PLCG2 mutations in 5 of 9 patients (55.6%) with detectable mutations (Figure 2C). Four patients harbored 2 to 3 different types of nonsynonymous mutations affecting different base-pair loci of either PLCG2 or BTK as exemplified in Figure 2D. The 6-nucleotide deletion of PLCG2 exon 20 coincided with BTK C481S mutations in 2 patients.
Emergence of resistance mutations during treatment
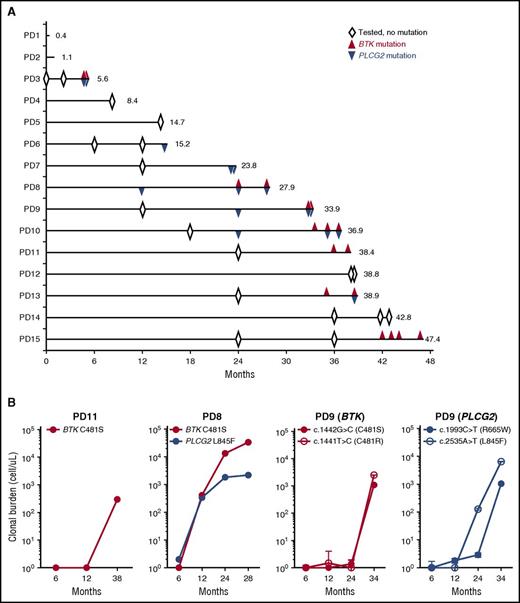
To test whether resistance-conferring mutations could be detected before manifestation of clinical progression, we performed targeted resequencing of stored samples obtained sequentially during the study (Figure 3A). Of 9 patients with BTK and/or PLCG2 mutations, 6 (66.7%) had detectable mutations predating clinical progression, often by many months (median, 8 months; range, 2.9-15.4 months) (supplemental Table 4). All patients except 1 had an early on-treatment sample available that tested negative for BTK and PLCG2 mutations, indicating expansion of subclones carrying drug-resistant mutations during treatment.
Clonal evolution during treatment with ibrutinib. (A) A swimmer plot of 15 patients who progressed on ibrutinib. Each lane represents an individual patient. Numbers at the end of the line indicate TTP (months). Open diamonds indicate BTK and PLCG2 candidate regions were tested for mutations, and no mutation was detected. Red triangles indicate the detection of BTK mutations. Blue triangles indicate PLCG2 mutations. (B) Growth kinetics of mutant clones. Three representative cases with BTK and/or PLCG2 mutations are shown (PD8, PD9, and PD11). Red color indicates BTK mutation. Blue color indicates PLCG2 mutation. The cell count of mutant clones was estimated from the VAF assessed by ddPCR (supplemental Table 2), the ALC, and the proportion of B cells (CD19+). Time points with VAFs below the lowest limit of detection (0.2%) are plotted as 1 cell (10°).
Clonal evolution during treatment with ibrutinib. (A) A swimmer plot of 15 patients who progressed on ibrutinib. Each lane represents an individual patient. Numbers at the end of the line indicate TTP (months). Open diamonds indicate BTK and PLCG2 candidate regions were tested for mutations, and no mutation was detected. Red triangles indicate the detection of BTK mutations. Blue triangles indicate PLCG2 mutations. (B) Growth kinetics of mutant clones. Three representative cases with BTK and/or PLCG2 mutations are shown (PD8, PD9, and PD11). Red color indicates BTK mutation. Blue color indicates PLCG2 mutation. The cell count of mutant clones was estimated from the VAF assessed by ddPCR (supplemental Table 2), the ALC, and the proportion of B cells (CD19+). Time points with VAFs below the lowest limit of detection (0.2%) are plotted as 1 cell (10°).
Next, we estimated the abundance and growth rate of the mutant subclones. VAFs were determined by ddPCR using CD19+ selected cells (supplemental Figure 2). We then calculated the absolute cell count of mutant subclones from the VAF and the proportion of CD19+ cells among the ALC. Figure 3B shows the emergence of different resistant subclones in 3 exemplary male patients. Patient PD11 progressed at 38.4 months with increasing lymphadenopathy. In the peripheral blood, 60.6% of the CD19+ cells carried a BTK C481S mutation, for an absolute mutant B-cell count of 300 cells/μL. The VAF at 6 and 12 months was below the limit of detection (<0.2%). In patient PD8, BTK and PLCG2 mutations simultaneously became first detectable at 12 months with VAFs of 2.5% and 1.0%, corresponding to clonal frequencies of 2.5% and 2.0%, respectively. The BTK-mutated clone expanded more rapidly than the PLCG2-mutated clone (estimated doubling times, 4.6 and 23.1 months, respectively), and became the dominant clone at progression with a clonal burden 15 times higher than the PLCG2-mutated clone (33 000 and 1100 cells/μL, respectively). Patient PD9 presented with multiple subclonal mutations that were first detectable at 24 months; at the time, 2 subclones with different PLCG2 mutations (R665W and L845F) were more abundant than the 2 subclones with BTK mutations (C481S and C481R). The 2 BTK mutant subclones expanded at a very similar (doubling times, 0.9-1.1 months) but faster rate than the PLCG2-mutant clones (doubling times, 2 months for L845F and 1.2 months for R665W). Based on the different growth rates of cells with PLCG2 mutations and BTK mutations, we concluded that the different mutations identify distinct subclones evolving independently.
Quality of treatment response and disease progression
The quality of response in patients developing secondary resistance appeared to be no different than in patients who have not progressed to date. Ten of 15 patients who later progressed achieved an objective response with ibrutinib, including 2 with complete responses (supplemental Table 4). Specifically, tumor control in peripheral blood and bone marrow as well as reductions in B2M were comparable in both PD and non-PD groups (supplemental Figure 3). Furthermore, the level of residual disease measured by peripheral blood flow cytometry at 12 and 24 months was not associated with the risk of relapse (supplemental Figure 4A-B). However, above median level of residual disease at 36 months was associated with an increased risk of disease progression over the subsequent 12 months (supplemental Figure 4C).
Biomarkers of treatment response and disease progression
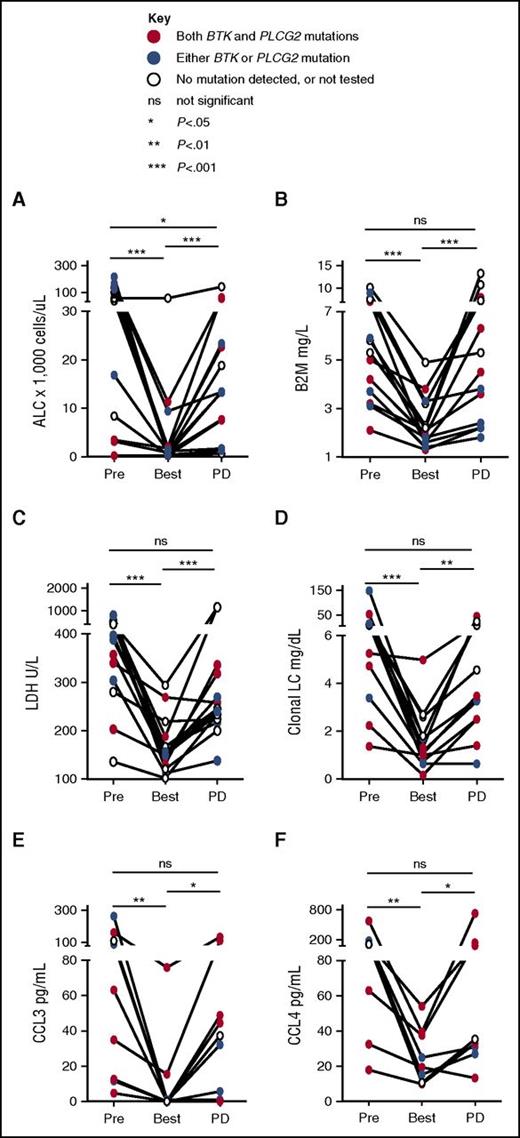
Next, we explored biomarkers of disease response at selected time points on study. ALC, B2M, lactate dehydrogenase (LDH), and clonal light chains significantly decreased from pretreatment baseline to best response (Figure 4A-D). At progression, the majority of patients showed a significant ALC increase compared with best response (median ALC, 13 400/μL at progression vs 1225/μL at best response; P < .001). Three patients, however, had only minimal changes in peripheral lymphocyte count at progression (ALC < 2000/μL). Across all patients, B2M, LDH, and clonal light chains significantly increased at progression (all P < .01). In the 3 patients with low ALC at progression, B2M increased by 1.1- to 1.4-fold, LDH by 1.2- to 2.3-fold, and clonal free light chain by 1.9- to 13.5-fold compared with best response.
Biomarkers of disease progression. ALC in panel A and serum-derived biomarkers were measured at baseline (Pre), best response (Best), and progression (PD) in matched samples: (B) B2M; (C) LDH; (D) clonal light chain (LC), either κ or λ, corresponding to the light chain restriction of the CLL cells as determined by flow cytometry; and BCR-regulated chemokines (E) CCL3; and (F) CCL4. Solid lines above each graph indicate results of Wilcoxon matched-pairs signed rank test. *P < .05, **P < .01, ***P < .001. ns, not statistically significant. PD, progression of disease.
Biomarkers of disease progression. ALC in panel A and serum-derived biomarkers were measured at baseline (Pre), best response (Best), and progression (PD) in matched samples: (B) B2M; (C) LDH; (D) clonal light chain (LC), either κ or λ, corresponding to the light chain restriction of the CLL cells as determined by flow cytometry; and BCR-regulated chemokines (E) CCL3; and (F) CCL4. Solid lines above each graph indicate results of Wilcoxon matched-pairs signed rank test. *P < .05, **P < .01, ***P < .001. ns, not statistically significant. PD, progression of disease.
CCL3 and CCL4 are chemokines secreted by CLL cells in response to BCR and NF-κB activation.30,31 Secretion of both chemokines is greatly decreased by ibrutinib in vitro and in vivo, providing a surrogate marker of on-target effects.5,32,33 Consistent with the clinical response, serum levels of CCL3 and CLL4 at the time of best response were significantly decreased compared with pretreatment (Figure 4E-F). In 6 of 9 patients who had available samples, serum levels of CCL3 and CLL4 rebounded at the time of clinical progression consistent with reactivation of BCR signaling in ibrutinib-resistant cells (Figure 4E-F).
Patient disposition and survival
Eight of the 15 patients died due to disease progression or complications related to salvage treatments (supplemental Table 5). Most patients who progressed with CLL received treatment with alternative targeted agents, and 7 were alive with the median estimated survival time from progression of 19.8 months (supplemental Figure 5). In contrast, all 5 patients with histologic transformation died within 8 months (median survival, 3.5 months; P = .005 for difference in survival).
Discussion
Targeting BCR signaling has changed the treatment paradigm for CLL.4 The BTK inhibitor ibrutinib is approved for the treatment of CLL both in first-line and for relapsed disease. Although single-agent therapy provides durable, well-tolerated disease control, resistance to ibrutinib is emerging as an area of unmet needs.17 Among 84 patients, 53 with TP53 aberration, enrolled on a phase 2 study at our institution, the rate of progression at the median follow-up of 3 years was 17.9%. Resistant disease was the most common reason for patients to discontinue treatment and was the leading cause of death.
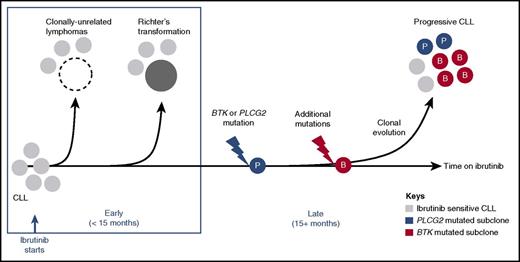
Transformation to aggressive disease accounted for most cases of early progression (Figure 5). In fact, all cases of disease transformation presented within the first 15 months. The absence of transformation on long-term therapy cannot be explained by observation bias as all cases with late progression had tissue biopsies performed to rule out transformation. Early manifestation of transformed disease suggests that the genetic events leading to transformation likely predate the start of treatment. The paucity of late transformation events in our study could be due to the limited sample size. However, given that the majority of patients had TP53 aberration, a high-risk disease subset at increased risk of transformation, an alternative explanation is that effective inhibition of BCR signaling could decrease the drive toward transformation. The latter view is consistent with an increased risk of transformation associated with stereotyped BCRs34 and the relatively high rates of Richter transformation in patients with TP53 aberration treated with FCR compared with ibrutinib-treated patients.11,35
Molecular patterns of ibrutinib-resistant disease. A schematic representation of study findings. Sensitivity to ibrutinib is altered by histology and the presence of resistance-conferring mutations. Histologic transformation is an early event (<15 months in the study presented here), and can be de novo (clonally unrelated) or originate in the CLL clone (clonally related). Progressive CLL occurs later in the treatment course, and is frequently accompanied by BTK (red circles) and/or PLCG2 (blue circles) mutations. Multiple subclones can coexist, precede clinical progression by many months, and expand at different rates.
Molecular patterns of ibrutinib-resistant disease. A schematic representation of study findings. Sensitivity to ibrutinib is altered by histology and the presence of resistance-conferring mutations. Histologic transformation is an early event (<15 months in the study presented here), and can be de novo (clonally unrelated) or originate in the CLL clone (clonally related). Progressive CLL occurs later in the treatment course, and is frequently accompanied by BTK (red circles) and/or PLCG2 (blue circles) mutations. Multiple subclones can coexist, precede clinical progression by many months, and expand at different rates.
In contrast to transformed disease, progressive CLL presented late, almost exclusively in patients treated for 2 or more years. In 6 of 10 patients with progressive CLL, increasing lymphadenopathy was the defining criterion. At progression, most patients showed an increase in B2M, LDH, and/or the clonal serum free light chain from baseline that, in the absence of confounding events, were quite useful as markers of disease activity. In this regard, it is important to consider that at the time of CLL progression disease burden was often minimal, adding value to noninvasive, nonradiologic tests suitable to sequential monitoring of patients over time.
BTK and/or PLCG2 mutations were present in 80.0% of patients progressing with CLL, confirming prior reports on the high prevalence of these mutations in patients with ibrutinib-resistant CLL.17 Most BTK mutations described here and previously by others, affect the cysteine residue to which ibrutinib covalently binds, leading to its replacement by a different amino acid: C481S, C481R, C481F, and C481Y.17,18,29 The diversity of substitute amino acids supports the notion that loss of the cysteine residue and consequently the loss of sustained target inhibition is indeed responsible for ibrutinib resistance. Consistently, the growth rate of 2 distinct subclones arising in the same patient, 1 with a C481S and the other with a C481R, were virtually identical (PD9; Figure 3B). Several nonsynonymous mutations in PLCG2 identified here have previously been reported in ibrutinib-resistant CLL including R665W, S707Y, and L845F.17,18,20 The P664S mutation and the 6-nucleotide deletion in exon 20 have not been previously described but are also located in the SH2 domain where the majority of PLCG2 mutations occur. Mutation and deletion of this autoinhibitory SH2 domain have been shown to induce BTK-independent activation of PLCγ2.19,36 Furthermore, R665W and L845F mutations were shown to increase sensitivity of PLCγ2 to Rac2 and reconstitute downstream BCR signaling independent of BTK activation.37
Ibrutinib-resistant CLL often harbored several subclonal mutations. Multiple subclones, 3 with unique mutations in BTK codon 481 and 2 with PLCG2 mutations, could be clearly identified in PD9. This conclusion is based on the demonstration of separate mutations in BTK on independent sequence strands aided by the X-chromosomal location of the gene. Furthermore, the different growth rates of the BTK and PLCG2 mutant subclones is most consistent with the mutations arising in independent ancestral cells. Similarly, the different growth rates of BTK- and PLCG2-mutant clones in PD8 again suggest that these are independent subclones evolving concurrently. However, we cannot exclude that in other patients multiple mutations coexist in the same cell, potentially even as biallelic mutations. Given that we only sequenced candidate regions for known resistance mutations, we likely underestimate the clonal complexity of ibrutinib-resistant disease. The subclonal heterogeneity of CLL progressing on ibrutinib has important clinical implications. For example, although reversible BTK inhibitors might be effective against BTK C481S-mutant clones, they would not be effective against PLCG2 mutants.
Prospective identification of patients at high risk for progression on single-agent ibrutinib may be useful to stratify patients to trials using combination therapy or alternative approaches such as venetoclax, immune checkpoint inhibitors, or chimeric antigen receptor–modified T cells.38 Del(17p) in relapsed/refractory patients and complex karyotype have been associated with a high risk of progression.12,39 In our series, del(17p) or TP53 mutation, advanced Rai stage, relapsed/refractory disease, and high B2M at study entry independently predicted an increased risk of progression. Among the 53 patients with TP53 aberration included in this study, 34 (64.2%) being treated in first-line, relapsed/refractory disease (HR, 2.67), advanced Rai stage (HR, 2.99), and elevated B2M (HR, 3.41) were associated with shorter PFS. G-banded karyotypes were not available in our patients as this test has been replaced by FISH cytogenetics.40
Using methods sensitive to ∼1 × 10−3, resistance mutations were detected as early as 15 months prior to clinical progression. In all patients with resistance mutations, an earlier on-treatment or pretreatment sample tested negative for the specific mutation, consistent with other reports on the absence of BTK and PLCG2 mutations at baseline.18,41 However, computational modeling suggested that mutations likely preexist at low frequency and are selected during treatment.42 Many assays may not be sensitive enough to detect such rare mutant cells. In fact, more recently, a PLCG2 mutation identified in the relapse sample was detected in 1 in 500 000 cells in the pretreatment sample using droplet-based PCR.20 Our observation that clones carrying BTK and PLCG2 resistance mutations had doubling times of ≤2 months, correlating to an ∼300 000-fold expansion of the resistant clone over 3 years is also consistent with the selection of rare resistant cells present at the initiation of treatment.
In most patients reported here, clinical observation prompted the submission of the first sample for sequencing. Upon detection of a mutation, stored samples from earlier time points were analyzed. In patients progressing after 2 years on ibrutinib, resistance mutations predated the clinical manifestation of progression often by many months. Patients with mutations who did not meet criteria for disease progression continued on ibrutinib under more frequent monitoring while alternative treatment options were being evaluated. Most patients who relapsed with CLL were able to successfully transition to salvage treatments consisting of phosphatidylinositol 3-kinase (PI3K) or Bcl-2 targeting agents, with median survival of 19.8 months. The survival of patients progressing with CLL in our series is comparable to that reported by the group at The Ohio State University with a median survival 17.6 months.17 The disease biology of late relapses and the availability of effective salvage agents likely account for the improvement in survival compared with early reports.16 In addition, seamless transition between treatments may forestall disease acceleration. It may, therefore, be important not to discontinue ibrutinib immediately when BTK and/or PLCG2 mutations are detected. Nevertheless, most patients with ibrutinib-resistant disease appear to eventually progress on salvage therapy and these patients should be considered early for allogeneic transplantation or chimeric antigen receptor–modified T-cell therapy. In contrast, for patients who progressed with transformation, subsequent therapy was largely ineffective and the median survival was short at 3.3 months. Further investigation is needed to identify effective therapeutic strategies for patients with Richter transformation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank their patients for participating and donating samples to make this research possible. The authors thank Susan Soto and Ajunae Wells for assistance in the clinic and Stephanie Housel, Adriana Byrnes, and Allison Wise for protocol support. The authors acknowledge Pharmacyclics for providing study drug and comments on a draft of this manuscript.
This work was supported by the Intramural Research Program of the National Institutes of Health National Heart, Lung, and Blood Institute and National Cancer Institute.
Authorship
Contribution: I.E.A. and A.W. conceived and designed the study; I.E.A., S.S., J.V., P.N., J.L., and M.F. conducted the clinical trial; I.E.A., C.U., A.A., S.E.M.H., I.M., D.C.A., L.W., S.P., C.M.Y., M.S.-S., L.X., M.R., M.F., and A.W. acquired data; I.E.A., C.U., X.T., M.A., and A.W. analyzed and interpreted data; A.W. supervised the study; and all authors wrote, reviewed, and revised the manuscript.
Conflict-of-interest disclosure: A.W. received research funding from Pharmacyclics Inc. The remaining authors declare no competing financial interests.
Correspondence: Adrian Wiestner, Hematology Branch, NHLBI, NIH, Building 10, CRC 3-5140, 10 Center Dr, Bethesda, MD 20892-1202; e-mail: wiestnera@mail.nih.gov.
References
Author notes
I.E.A.and C.U. contributed equally to this work.

![Figure 1. PFS. Kaplan-Meier estimates of PFS of (A) all patients and subgroups stratified by: (B) TP53 aberration (absent vs present), (C) prior treatment (RR CLL vs TN), (D) Rai stage (I/II vs III/IV), (E) B2M (≤4 mg/L vs >4 mg/L), and (F) IGHV mutational status (mutated IGHV [M] vs unmutated IGHV [U]). Among patients with TP53 aberration (n = 53), subgroups are further divided by: (G) prior treatment (RR vs TN), (H) Rai stage (I/II vs III/IV), and (I) B2M (≤4 mg/L vs >4 mg/L).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/129/11/10.1182_blood-2016-06-719294/4/m_blood719294f1.jpeg?Expires=1769147724&Signature=srYHshhU7eJJG-35Qdb2glXUkLChp2rstTxtsy2eNRS57UTNknAyVoy2dg8swhRaR3yjEkMvbih8Wu8yEPL42-TelKMrPFDwMUIbWInrW5LW5wxRsZHKUWsiRQDez9y05NeKAcqS5cpfPJmMLq3TYdm7wi5Bk6K9BPMUhjvoPi3z~L23kjezbBOIzLnvO~JutaGzNdZ-PDDzwCm~Vis~kNvA~2r0zsJRU1D-woqRgDPLoHxa-po60UZo3Lnp7pQc-iG4UpD8m7JU4AKkt-5duO6LnVCAq1e72zSIFVm2ifz201bqirkJRNOVPBFhF0Iw13Dkhh2-5TlH2~nK-RLcdw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)