Key Points
A pharmacological screening identified compounds that reactivate B-cell–specific gene expression in cHL cell lines.
B-cell phenotype-restoring drug combinations render cHL cell lines susceptible to B-NHL–reminiscent targeted therapies.
Abstract
Classical Hodgkin lymphoma (cHL), although originating from B cells, is characterized by the virtual lack of gene products whose expression constitutes the B-cell phenotype. Epigenetic repression of B-cell–specific genes via promoter hypermethylation and histone deacetylation as well as compromised expression of B-cell–committed transcription factors were previously reported to contribute to the lost B-cell phenotype in cHL. Restoring the B-cell phenotype may not only correct a central malignant property, but it may also render cHL susceptible to clinically established antibody therapies targeting B-cell surface receptors or small compounds interfering with B-cell receptor signaling. We conducted a high-throughput pharmacological screening based on >28 000 compounds in cHL cell lines carrying a CD19 reporter to identify drugs that promote reexpression of the B-cell phenotype. Three chemicals were retrieved that robustly enhanced CD19 transcription. Subsequent chromatin immunoprecipitation-based analyses indicated that action of 2 of these compounds was associated with lowered levels of the transcriptionally repressive lysine 9-trimethylated histone H3 mark at the CD19 promoter. Moreover, the antileukemia agents all-trans retinoic acid and arsenic trioxide (ATO) were found to reconstitute the silenced B-cell transcriptional program and reduce viability of cHL cell lines. When applied in combination with a screening-identified chemical, ATO evoked reexpression of the CD20 antigen, which could be further therapeutically exploited by enabling CD20 antibody-mediated apoptosis of cHL cells. Furthermore, restoration of the B-cell phenotype also rendered cHL cells susceptible to the B-cell non-Hodgkin lymphoma-tailored small-compound inhibitors ibrutinib and idelalisib. In essence, we report here a conceptually novel, redifferentiation-based treatment strategy for cHL.
Introduction
Classical Hodgkin lymphoma (cHL), although a treatable malignancy with a high likelihood of cure for most patients, reflects a clinical challenge when presenting as primary refractory or relapsed disease. Interestingly, cHL is a paradigm example of malignant plasticity,1 which accounts for the limited effectiveness of modern B-cell–specific targeted therapeutics in this entity. cHL originates from B cells because the malignant Hodgkin-Reed-Sternberg (HRS) cells harbor rearrangements and mutations due to somatic hypermutation of their immunoglobulin gene loci.2,3 However, unlike B-cell non-Hodgkin lymphomas (B-NHLs), cHL cells usually lack expression of B-cell–specific gene products such as CD19, CD20, the B-cell receptor (BCR), and associated components like CD79a and CD79b.4 Previous studies showed that downregulation of octamer-dependent transcription factor 2 (Oct2), its coactivator BOB.1, and the transcription factor PU.1 contribute to this phenotypic transformation in cHL.5,6 The early B-cell differentiation factors, namely E2A, EBF, PAX5, and also FoxO1, are either downregulated and often hardly detectable in HRS cells,3,7,8 or functionally compromised by aberrantly high expression levels of B-cell–inappropriate transcription regulators like ID2, ABF-1, and NOTCH1.9-11 In addition to deregulated transcription factor networks, epigenetic alterations were observed in cHL: for instance, the promoter region of the IgH locus was found to be decorated with the transcriptionally repressive lysine 9-trimethylated histone H3 mark (H3K9me3) in the cHL cell lines L428 and L1236.12 Whether DNA hypermethylation of B-cell–relevant gene promoters (eg, at the PU.1, BOB.1, CD19, and CD79B loci) critically contributes to the lost B-cell phenotype in primary HRS cells and cHL cell lines, compared with normal B cells, remains a controversy in the field.13-15 Because impaired expression of B-cell markers may promote aggressive tumor growth at the cell-autonomous level, and, possibly, by interfering with antilymphoma immunosurveillance,16 restoration of the B-cell phenotype might also be of therapeutic benefit in cHL.
Moreover, antibodies against B-cell surface receptors, especially against the CD20 antigen, have fundamentally changed the clinical outcome of B-NHL patients.17 In addition to previously registered CD20 antibodies such as rituximab and tositumomab, CD19, for example, is being targeted by blinatumomab, a bispecific anti-CD19/CD3 T-cell–engaging antibody, or CD19-recognizing chimeric antigen receptor T cells, and antibody-drug conjugates are currently being tested against B-cell surface receptors such as CD22 and CD79.18-21 Aiming to reinduce the silenced B-cell program, particularly CD19, CD20, and CD79 surface receptors in cHL cells, we carried out large-scale pharmacological screening in stably CD19 promoter reporter-engineered cHL cells, further explored promising compounds as well as the differentiation-inducing agents all-trans retinoic acid (ATRA) and arsenic trioxide (ATO) in functional assays, and specifically tested their resensitization potential for CD20-directed or BCR signaling targeting cotherapies.
Materials and methods
Lymphoma tissue sections
The anonymous use of human lymphoma biopsies primarily obtained for the initial diagnosis was based on informed patient consent, and approved by the local ethics commission (reference EA4/104/11). Immunohistochemical stainings of formalin-fixed and paraffin-embedded tissue sections were carried out as described.22 Antibody information is provided in supplemental Table 1 (available on the Blood Web site). Images were acquired using Diskus software (Hilgers Technisches Büro, Königswinter, Germany) with a Leica microscope DM RXA using a 40× objective (Leica Microsystems, Wetzlar, Germany) and a JVC camera, model KY-F75U (Yokohama, Japan).
Cell culture
The human cell lines KM-H2, L428, L540, L591, L1236, SUP-HD1, U-HO1, and HDLM-2 (all cHL), SU-DHL4, SU-DHL5, SU-DHL10, BL60, Daudi, Karpas 422, and Namalwa (all B-NHL) and Nalm6 (pre-B acute lymphoblastic leukemia) were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum.
Plasmid construction
A full-length complementary DNA of the human bcl2 gene was cloned into a retroviral murine stem cell virus (MSCV) backbone with a blasticidin resistance gene, and a mifepristone-inducible human PAX5 expression system (GeneSwitch System; Life Technologies/Invitrogen, Carlsbad, CA) was generated as described.23,24 Plasmids encoding B-cell–specific promoter sequences and a geneticin (G418) resistance gene were generous gifts from C. Tonnelle (EμMar-CD19-pTRIP) and M. Sigvardsson (pGL3-mb-1 [CD79a] and pGL3-B29 [CD79b]).25-27 The EμMar-CD19 fragment was further subcloned into the luciferase-encoding pGL3 reporter vector system as well. Gene-specific small hairpins against the histone-lysine N-methyltransferase EHMT2 were cloned into the pLKO1 lentivirus. Short hairpin RNA sequence and TaqMan assay information are provided in supplemental Table 2.
Transduction procedures
Stable infection of cHL and B-NHL cells with MSCV-based retroviruses was carried out after transduction with an ecotropic receptor-encoding plasmid as described.23 pGL3-based promoter reporter constructs and the PAX5-overexpressing plasmid were delivered via nucleofection (Lonza). Bcl2- and PAX5-overexpressing cHL cells were only used in the screening, that is referring to results in Figure 2A-C. The pmaxGFP plasmid was transfected to monitor transduction efficacy and viability of the target cells. Stable integrants were clonally expanded from single cells by G418 (Sigma-Aldrich) treatment, and verified by polymerase chain reaction (PCR) using pGL3-specific primers. A list of primer sequences is provided in supplemental Table 2.
Luciferase reporter assays
To detect reporter activity, 50 μL of One-Glo reagent (Promega) was added to 50 μL of cell suspension, and incubated at room temperature for 15 minutes. For normalization, 10 μM calcein (CAM; Sigma-Aldrich) was added 10 minutes prior to the measurement of fluorescence signals from live cells.
Flow cytometric analyses
Cells were washed and incubated with directly conjugated primary antibodies for 30 minutes on ice, and analyzed using a Becton Dickinson FACSCalibur. Immunofluorescence image-based flow cytometry was conducted on an Amnis ImageStreamX flow cytometer. As a negative control, the corresponding isotype control antibody was used, and mean fluorescence intensity (MFI) values were calculated by subtracting the MFI of the isotype control. Antibody information is provided in supplemental Table 1.
Pharmacological compound library screening and other drugs used
The pharmacological library screen was performed in 384-well plates using a library of 28 160 chemical compounds, enriched for potentially bioactive agents (ChemBioNet).28 Drugs were added at a final concentration of 10-µM to 40-µL cell suspension. The One-Glo kit (Promega) was used to detect the luciferase-based reporter signal. After 48 hours of incubation time, signals were captured using a TECAN microplate reader. Data were analyzed based on the Z score (the number of standard deviations [SDs] a measured signal intensity is above the mean) and the Tanimoto score (indicating the extent of similarity between 2 molecules), referring to the “Functional Class FingerPrints of maximum diameter_4 (FCFP_4)” algorithm.29 The 3 newly identified compounds 27 (R367-0003 from ChemDiv), 40 (CTK6G9834 from ChemTik and Vitas-M Laboratory), and 49 (BAS 05262891 from Asinex) were dissolved in 10 mM dimethyl sulfoxide. Information on additional drugs (and their solvents) is provided in supplemental Table 3.
Gene expression analyses
Total RNA was prepared using the TRIzol reagent (Invitrogen). Oligo(dT)-primed complementary DNA was synthesized with the Superscript II Reverse Transcriptase (Invitrogen). Quantitative reverse transcription PCR (RQ-PCR) analysis was performed using the TaqMan Gene Expression assay (Applied Biosystems) with the housekeeping gene succinate dehydrogenase complex, subunit A as an internal control. All reactions were performed in triplicates. Relative transcripts levels were calculated based on the comparative ΔΔ cycle threshold method.30 A list of TaqMan assays used is available in supplemental Table 2. Immunoblot analyses were conducted as described.30 Densitometric analyses of some immunoblots were carried out using the ImageJ software package. Antibody information is available in supplemental Table 1.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed using the ChIP-IT Express kit (Active Motif), with antibodies specific to H3K4me3, H3K9me3, and H3K27me3. Immunoprecipitated DNA was amplified by real-time PCR, and the signals were normalized to input DNA. Antibody information and primer sequences can be found in supplemental Tables 1-2.
Viability assay
The Guava ViaCount assay was performed using a Guava easyCyte flow cytometer and the Guava CytoSoft software package (Millipore). After staining of the cells with ViaCount reagent, viable and dead cells were separated using the viability (PM1) vs nucleated cells (PM2) plot.
Antibody-dependent cellular cytotoxicity
Antibody-dependent cellular cytotoxicity (ADCC) was assessed using the nuclear factor of activated T cells (NFAT)-driven luciferase ADCC Reporter Bioassay kit (Promega). After pretreatment of the cells with the indicated agents as stated in the respective legend or left untreated as a control, rituximab (at 20 µg/mL; without cross-linker) or tositumomab (at 10 µg/mL) was added for 1 hour, followed by the NFAT-luciferase–engineered effector Jurkat T cells. Signal intensities were calculated as values relative to the signal from untreated cells without the anti-CD20 antibody.
Statistical analysis
The unpaired Student t test was used to compare means and SDs or standard deviations of the mean (SEM) as indicated. A P value <.05 was considered statistically significant and was marked by an asterisk.
Results
Monitoring of the B-cell–specific transcription program in cHL and B-NHL reporter cell lines
To confirm loss of the B-cell phenotype, as demonstrated in cHL patient biopsies (Figure 1A) in cHL cells used in subsequent investigations here, we first immunophenotyped cHL and B-NHL cell lines regarding a variety of B-cell– and cHL-typical surface markers. Flow cytometric analyses demonstrated the virtual absence of CD19, CD20, and CD79b expression in the CD15+ and CD30+ cHL cell lines, contrasting opposite findings in B-NHL cell lines (Figure 1B; supplemental Figure 1). To sensitively monitor and visualize B-cell–characteristic gene expression, we generated cHL and B-NHL cell line clones harboring luciferase reporter constructs under control of the B-cell–specific CD19, CD79a (mb-1), or CD79b (B29) promoters (supplemental Figure 2A).
Modulation of CD19 promoter activity in cHL and B-NHL cell lines. (A) Expression of the B-cell–specific surface marker CD20 on B-NHL samples (follicular lymphoma [FL], left; diffuse large B-cell lymphoma [DLBCL], right), but not on cHL samples (with CD30 as a typical cHL marker). Standard hematoxylin and eosin (H.E.) staining to visualize morphology. Note CD20− HRS (arrowheads) surrounded by a few infiltrating, CD20+ nonmalignant B cells in the cHL sections. (B) Endogenous CD19 surface antigen expression levels detected by flow cytometry. MFI of individual cHL and B-NHL cell lines (left), and group average of each entity (right). (C) Firefly luciferase-detected CD19 promoter reporter activity, normalized by total protein content in a similar panel of individual (left) and entity-grouped (right) cHL and B-NHL cell lines as in panel B. (D) MFI of CD19 surface antigen expression in cHL and B-NHL cell lines upon 5-Aza/TSA treatment or left untreated (UT) by flow cytometry (individual cell lines, left; average of B-NHL cell lines, right). (E) Firefly luciferase–indicated CD19 promoter activity in cHL and B-NHL cell lines upon 5-Aza/TSA treatment (as in panel D). Data are presented as mean ± SEM. All experiments were done at least in triplicate; *P < .05 throughout the figure.
Modulation of CD19 promoter activity in cHL and B-NHL cell lines. (A) Expression of the B-cell–specific surface marker CD20 on B-NHL samples (follicular lymphoma [FL], left; diffuse large B-cell lymphoma [DLBCL], right), but not on cHL samples (with CD30 as a typical cHL marker). Standard hematoxylin and eosin (H.E.) staining to visualize morphology. Note CD20− HRS (arrowheads) surrounded by a few infiltrating, CD20+ nonmalignant B cells in the cHL sections. (B) Endogenous CD19 surface antigen expression levels detected by flow cytometry. MFI of individual cHL and B-NHL cell lines (left), and group average of each entity (right). (C) Firefly luciferase-detected CD19 promoter reporter activity, normalized by total protein content in a similar panel of individual (left) and entity-grouped (right) cHL and B-NHL cell lines as in panel B. (D) MFI of CD19 surface antigen expression in cHL and B-NHL cell lines upon 5-Aza/TSA treatment or left untreated (UT) by flow cytometry (individual cell lines, left; average of B-NHL cell lines, right). (E) Firefly luciferase–indicated CD19 promoter activity in cHL and B-NHL cell lines upon 5-Aza/TSA treatment (as in panel D). Data are presented as mean ± SEM. All experiments were done at least in triplicate; *P < .05 throughout the figure.
Consistent with the loss of B-cell–specific gene products, we found dramatically lower CD19 promoter activities in all cHL cell lines tested when compared with B-NHL cell lines (Figure 1C). Because activity signals of CD79a and CD79b promoter-driven constructs were not overtly different (supplemental Figure 2B-C), we decided to focus on the subsequent screening on the CD19 reporter system as readout.
To explain the lost B-cell phenotype, one might hypothesize silencing of the respective transcriptional networks in cHL. However, the DNA-demethylating agent 5-aza-2′-deoxycytidine (5-Aza) alone, or even more profoundly in combination with the histone deacetylase inhibitor trichostatin A (TSA), reportedly reduced CD19 transcript levels in B-NHL cells,15 suggesting that a CD19-suppressive factor, a panel of negative regulators, or even a singular master repressor of the B-cell program was epigenetically inactivated in B-NHL, while possibly remaining active in cHL. We confirmed and extended this finding by flow cytometry, which demonstrated reduced membrane expression of CD19 protein in 5-Aza- or 5-Aza/TSA-exposed B-NHL, and no reexpression in equally treated cHL cell lines (Figure 1D). Likewise, these treatments resulted in much lower CD19 promoter activities exclusively in B-NHL reporter cell lines (Figure 1E).
A high-throughput screening identifies compounds that restore CD19 transcription in cHL cell lines
Having this CD19 reporter system established, we next set up a pharmacological screening to identify compounds that reinduce the lost B-cell phenotype in cHL cells. First, we stably transduced cHL cell lines with bcl2 to protect them against potential proapoptotic pressure, which may arise from B-cell–specific gene reexpression. Second, PAX5, a transcription factor critical for B-cell commitment, is often moderately expressed in HRS cells irrespective of their lost B-cell phenotype31 but virtually undetectable in cHL cell lines KM-H2 and L428 (supplemental Figure 3A). To rule out that subcritical PAX5 expression may hinder reconstitution of CD19 promoter activity, we infected L428 cells prior to the pharmacological screening with a mifepristone-inducible construct encoding PAX524 (supplemental Figure 3B).
Using these PAX5/Bcl2-engineered L428 reporter cells, we conducted a luciferase-based pharmacological screening of a 28 160-compound library enriched for potentially bioactive compounds. After vigorous selection, we finally came up with a list of 55 compounds (supplemental Figure 3C-D). We treated the 3 bcl2-infected cHL reporter cell lines L428, L1236, and KM-H2 (all without exogenous PAX5) with all 55 compounds, and selected 13 chemicals (reflecting 6 structurally distinct groups) that were effective in all 3 cHL lines and possessed good structure soundness and derivation possibility (Figure 2A). As a last selection step, we tested these 13 compounds regarding their ability to drive endogenous CD19 expression in nonengineered cHL cell lines, leaving us with 3 compounds: compounds encoded as “27” and “49” increased CD19 expression in L428 and KM-H2 cells, whereas compound “40” induced CD19 transcripts in L1236 cells (Figure 2B). Structurally, compounds 27, 40, and 49 share limited similarity among each other (Figure 2C). Chemically, compound 27 (PubChem database ID: 732887) is ethyl-3-amino-1H-indole-2-carboxylate, compound 40 (ChEMBL database ID: CHEMBL1457311) 2-(4-chlorophenyl)-1H-benzimidazol-5-amine, and compound 49 (ID: CHEMBL1424463) 1-(5-[(4-methoxyphenyl)amino]-1,2,4-thiadiazol-3-yl)-acetone. Although the pharmacological mode of action remains to be elucidated with respect to compound 27, compounds 40 and 49 have been previously claimed to interfere with epigenetic regulators32-34 (see next section and Discussion for details), a suggestive and attractive mechanism by which they may contribute to the reexpression of a silenced B-cell program.
A pharmacological library screen leading to the identification of 3 compounds that stimulate CD19 gene transcription in cHL cells. (A) Firefly luciferase–based CD19 reporter readout validation of 55 preselected compounds in 3 cHL reporter cell lines. Cells were incubated with the respective chemicals at 10 μM for 48 hours or left untreated (UT). Fold-induction values >2 are marked in red (see color coding). Numeric codes of the compounds are listed at the bottom, highlighted in yellow or blank groups indicating similar structure. Blue boxes mark 6 groups of chemicals, comprising 13 compounds (see supplemental Figure 3D), as effective in all 3 cHL cell lines. (B) Stimulation of CD19 transcription in cHL cell lines after a 48-hour exposure to 10 μM of compound 27, 40, and 49 (as marked in panel A), detected by RQ-PCR (presented as mean ± SD; *P < .05). (C) Structural formulas of screen-identified chemical compounds 27, 40, and 49.
A pharmacological library screen leading to the identification of 3 compounds that stimulate CD19 gene transcription in cHL cells. (A) Firefly luciferase–based CD19 reporter readout validation of 55 preselected compounds in 3 cHL reporter cell lines. Cells were incubated with the respective chemicals at 10 μM for 48 hours or left untreated (UT). Fold-induction values >2 are marked in red (see color coding). Numeric codes of the compounds are listed at the bottom, highlighted in yellow or blank groups indicating similar structure. Blue boxes mark 6 groups of chemicals, comprising 13 compounds (see supplemental Figure 3D), as effective in all 3 cHL cell lines. (B) Stimulation of CD19 transcription in cHL cell lines after a 48-hour exposure to 10 μM of compound 27, 40, and 49 (as marked in panel A), detected by RQ-PCR (presented as mean ± SD; *P < .05). (C) Structural formulas of screen-identified chemical compounds 27, 40, and 49.
Pharmacological rescue of the B-cell phenotype is associated with histone H3 modifications
Previous studies observed histone H3 hypermethylation in the promoter region of B-cell–specific genes in cHL cell lines.12,15 We performed ChIP assays to analyze a variety of histone modifications, namely histone H3K9me3 and H3K27me3 as transcriptionally repressive and H3K4me3 as rather transcription-promoting chromatin marks, at the CD19 promoter (of note, in cHL cells not being engineered with exogenous promoter/reporter constructs). Five pairs of primers (P1-P5) were designed to amplify a region spanning 1000 bp upstream to 200 bp downstream of the transcription start site. Compared with the untreated condition, exposure to compound 49, which produced a 48-fold increase of CD19 transcript levels in L428 cells (compare Figure 2B), led to markedly reduced H3K9me3 and H3K27me3 occupation at the CD19 promoter in L428 cells (Figure 3A-B). Treatment of L1236 cells with compound 40, enhancing CD19 transcript expression by about sixfold (compare Figure 2B), resulted in downregulated H3K9me3 and upregulated H3K4me3 marks at the CD19 promoter (Figure 3C-D). In line with a presumably nonepigenetic mode of its pharmacological action, we did not observe any changes in CD19 promoter occupation according to the 3 ChIP analyses (carried out in L428 cells) in response to compound 27 that would explain the CD19 transcript-inducing activity of this agent (data not shown). In essence, the data regarding the 2 “epigenetic” compounds 40 and 49 are suggestive of a certain pattern: the conversion of heterochromatinized regions (ie, especially P1/2 and P4/5) of the CD19 promoter into a more euchromatin-characteristic state that becomes permissive for the B-cell–specific transcription factor machinery to drive gene expression.
Candidate compound-related epigenetic changes at distinct histone H3 methylation sites around the CD19 promoter. (A-B) Relative levels of H3K9me3 (A) and H3K27me3 (B) in the CD19 promoter regions P1-P5 of compound 49–exposed L428 cells, detected by ChIP, normalized to the untreated control (UT). (C-D) Relative levels (as in panels A and B) of H3K9me3 (C) and H3K4me3 (D) in compound 40–exposed L1236 cells. Cells were treated with each compound at 10 μM for 48 hours. Data are presented as mean ± SD; *P < .05.
Candidate compound-related epigenetic changes at distinct histone H3 methylation sites around the CD19 promoter. (A-B) Relative levels of H3K9me3 (A) and H3K27me3 (B) in the CD19 promoter regions P1-P5 of compound 49–exposed L428 cells, detected by ChIP, normalized to the untreated control (UT). (C-D) Relative levels (as in panels A and B) of H3K9me3 (C) and H3K4me3 (D) in compound 40–exposed L1236 cells. Cells were treated with each compound at 10 μM for 48 hours. Data are presented as mean ± SD; *P < .05.
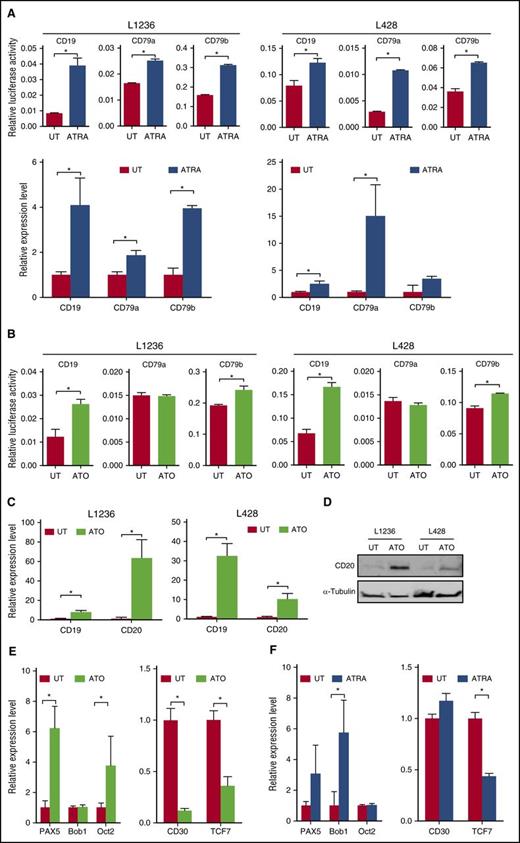
ATRA or ATO treatment initiates B-cell–specific transcription in cHL cell lines
To stress the idea of pharmacological restorability of the lost B-cell phenotype in cHL cells even further, we considered ATRA (not part of the drug library screened here) an attractive candidate. ATRA, the agent that overcomes the pathognomonic differentiation block in t(15;17)+ acute promyelocytic leukemia, was previously reported to promote expansion of CD19+ B cells in preclinical models, although the underlying mechanism, proliferation, differentiation, or selective induction of CD19 gene expression, was not unveiled.35 To explore whether ATRA may reinduce the B-cell program in cHL cells, we measured a variety of B-cell–specific promoter activities in cHL cell lines (without exogenous PAX5 restoration) in response to ATRA. L428 and L1236 cells consistently exhibited enhanced CD19, CD79a, and CD79b promoter activities, and expressed, accordingly, increased transcript levels of these B-cell markers after ATRA (Figure 4A). Interestingly, the reduction of CD19 expression in B-NHL cells after 5-Aza/TSA double treatment (compare Figure 1D-E) was completely neutralized if ATRA was added to the combination (supplemental Figure 4). This observation suggests that the putative repressor of the B-cell program, which is presumably silenced in B-NHL and normal B cells but active in cHL cells, cannot exert its repressive activity in the presence of ATRA.
Reconstitution of B-cell–specific gene expression by ATRA/ATO in cHL cell lines. (A) Stimulation of CD19, CD79a, and CD79b transcription by ATRA in L1236 (left) and L428 (right) cell lines detected by firefly luciferase reporter activity (top) and RQ-PCR (bottom). The luciferase signal was normalized to the cell number measured via calcein-generated fluorescence. Cells were treated with ATRA at 10 μM for 48 hours or left untreated (UT). (B) As in panel A, but treated with ATO at 10 μM for 48 hours. (C) CD19 and CD20 transcript levels in response to ATO in L1236 (left) and L428 (right) cell lines (as in panel B), detected by RQ-PCR. (D) Immunoblot analysis of CD20 protein expression in cHL cell lines as in panel B. α-Tubulin serves as a loading control. (E) Expression of B-cell–related transcription factor transcripts (left) and Hodgkin-typical transcripts (right) after ATO treatment in L1236 cells (as in panel B). (F) Expression of transcripts in L1236 cells as in E, but in response to ATRA (10 µM for 48 hours). Data are presented as mean ± SD; *P < .05.
Reconstitution of B-cell–specific gene expression by ATRA/ATO in cHL cell lines. (A) Stimulation of CD19, CD79a, and CD79b transcription by ATRA in L1236 (left) and L428 (right) cell lines detected by firefly luciferase reporter activity (top) and RQ-PCR (bottom). The luciferase signal was normalized to the cell number measured via calcein-generated fluorescence. Cells were treated with ATRA at 10 μM for 48 hours or left untreated (UT). (B) As in panel A, but treated with ATO at 10 μM for 48 hours. (C) CD19 and CD20 transcript levels in response to ATO in L1236 (left) and L428 (right) cell lines (as in panel B), detected by RQ-PCR. (D) Immunoblot analysis of CD20 protein expression in cHL cell lines as in panel B. α-Tubulin serves as a loading control. (E) Expression of B-cell–related transcription factor transcripts (left) and Hodgkin-typical transcripts (right) after ATO treatment in L1236 cells (as in panel B). (F) Expression of transcripts in L1236 cells as in E, but in response to ATRA (10 µM for 48 hours). Data are presented as mean ± SD; *P < .05.
ATO (not part of the screened library either), another agent used to treat acute promyelocytic leukemia patients, reportedly inhibited the constitutive NF-κB activity in cHL cells,36 which might be indirectly related to potential effects of this compound regarding the B-cell phenotype we seek to uncover here. Therefore, we treated L1236 and L428 cells with ATO, and analyzed its effects on B-cell–specific gene expression. Expression of CD19 and CD20 transcripts was markedly upregulated in both cell lines, even exceeding the effects seen in response to ATRA at its given dose schedule (Figure 4B-C). Of note, endogenous derepressed CD19 expression levels in cHL cells after exposure to a “restoring” agent, despite their profound elevation from virtual nondetectability, were still lower by orders of magnitude when compared with CD19 expression in untreated B-NHL cells (supplemental Figure 5). Importantly, we also detected strong CD20-protein expression signals when we probed cHL cell lines L1236 and L428 by immunoblot analysis (Figure 4D). ATO and ATRA enhanced transcript levels of the B-cell–specific transcription factors PAX5 and Oct2 or BOB.1, respectively, whereas the expression levels of the Hodgkin-typical gene product CD30 in response to ATO as well as the cHL-reminiscent and lineage-inappropriate TCF7 transcript in response to either agent were strongly reduced in L1236 cells (Figure 4E-F).5,31,37 Taken together, ATO and ATRA appear to “fix” the aberrant lineage fidelity in cHL cells by reinducing B-cell–specific transcription factors and B-cell–typical differentiation markers, and by repressing gene products indicative of the Hodgkin-reminiscent transdifferentiation phenotype.
Compound 40 enhances ATO-licensed anti-CD20-induced cell death in cHL cell lines
Consistent with previous reports on cytotoxic effects exerted by high-dose ATRA in cHL cell lines,38-40 reestablishment of the B-cell program in cHL cells by single-agent ATO or ATRA treatment resulted in a robust dose-dependent reduction of cell viability, suggestive of a prosurvival role the lost B-cell phenotype may have in Hodgkin biology (Figure 5A-B). Because ATO stimulated reexpression of CD20 in cHL cells (compare Figure 4D), we speculated whether it may, in addition, create the molecular basis for de novo sensitivity to a therapeutic CD20 antibody approach. L1236 cells were exposed to ATO or ATRA for 24 hours to induce CD20 expression, followed by incubation with the CD20 antibodies rituximab or tositumomab for 3 days prior to assessing cell viability. Compared with control samples that were treated with antibody but no ATO or ATRA, tositumomab-targeted L1236 cells presented with significantly lower viability when preexposed to a relatively low dose of ATO (10 μM) or ATRA (40 μM), with the latter agent also enhancing rituximab-induced cytotoxicity (Figure 5C). Rituximab or tositumomab alone, as anticipated, had no effect against the originally CD20− cHL cells. Of note, the kinetics of antibody-induced cell death are expectedly rather slow and less pronounced, if the actual contribution of host immunity in vivo, that is, ADCC and complement-mediated cytolysis, is not covered by the experimental setup. Moreover, an in vitro cytotoxicity assay may underestimate the activity of rituximab because cellular senescence, a terminal cell-cycle arrest, has just been unveiled as part of its antilymphoma action.41
Anti-CD20 cytotoxicity after single-agent or combination pretreatment. (A) Relative viability of L1236 cells after exposure to ATO at the indicated concentrations for 48 hours, normalized to the untreated control (UT). (B) As in panel A, but after ATRA. (C) Relative viability of L1236 cells pretreated with 10 µM ATO, 40 µM ATRA, or dimethyl sulfoxide (UT), subsequently exposed to CD20 antibodies rituximab (20 µg/mL) or tositumomab (10 µg/mL) or no antibody for 3 days. Percentages reflect normalization to the no-treatment setting. (D) CD20 expression after single-agent and combination treatments (as indicated) in L1236 cells, at the transcript level (top; relative values by RQ-PCR, normalized to the untreated control [UT]), and at the protein level (bottom; by immunoblot analysis, with α-tubulin as a loading control). ATRA, 20 μM; ATO, 5 μM; compound 40, 10 μM. (E) CD20 surface expression by immunofluorescence image-based flow cytometry (bottom) and matching brightfield pictures (top) in untreated (UT) SU-DHL4 B-NHL cells and L1236 cells exposed to ATO/compound 40 as in panel D (ATO/40), or untreated (UT); shown are 2 representative examples each. Of note, the L1236 signals are displayed with a fourfold amplification compared with the SU-DHL4 signals. (F) CD20 surface expression by conventional flow cytometry in L1236 cells as in panel E. (G) Relative viability of L1236 cells exposed to the CD20 antibodies rituximab or tositumomab (as in panel C) after the indicated pretreatments (as in panel D, ie, ATO at only 5 µM). (H) Rituximab-mediated ADCC values in L1236 (left) or L428 cHL cells (right) preexposed to the indicated drugs or combinations (as in panel G), 6 hours after the addition of the NFAT-luciferase–engineered effector T cells, relative to no preexposure (UT). Data are presented as mean ± SD; *P < .05.
Anti-CD20 cytotoxicity after single-agent or combination pretreatment. (A) Relative viability of L1236 cells after exposure to ATO at the indicated concentrations for 48 hours, normalized to the untreated control (UT). (B) As in panel A, but after ATRA. (C) Relative viability of L1236 cells pretreated with 10 µM ATO, 40 µM ATRA, or dimethyl sulfoxide (UT), subsequently exposed to CD20 antibodies rituximab (20 µg/mL) or tositumomab (10 µg/mL) or no antibody for 3 days. Percentages reflect normalization to the no-treatment setting. (D) CD20 expression after single-agent and combination treatments (as indicated) in L1236 cells, at the transcript level (top; relative values by RQ-PCR, normalized to the untreated control [UT]), and at the protein level (bottom; by immunoblot analysis, with α-tubulin as a loading control). ATRA, 20 μM; ATO, 5 μM; compound 40, 10 μM. (E) CD20 surface expression by immunofluorescence image-based flow cytometry (bottom) and matching brightfield pictures (top) in untreated (UT) SU-DHL4 B-NHL cells and L1236 cells exposed to ATO/compound 40 as in panel D (ATO/40), or untreated (UT); shown are 2 representative examples each. Of note, the L1236 signals are displayed with a fourfold amplification compared with the SU-DHL4 signals. (F) CD20 surface expression by conventional flow cytometry in L1236 cells as in panel E. (G) Relative viability of L1236 cells exposed to the CD20 antibodies rituximab or tositumomab (as in panel C) after the indicated pretreatments (as in panel D, ie, ATO at only 5 µM). (H) Rituximab-mediated ADCC values in L1236 (left) or L428 cHL cells (right) preexposed to the indicated drugs or combinations (as in panel G), 6 hours after the addition of the NFAT-luciferase–engineered effector T cells, relative to no preexposure (UT). Data are presented as mean ± SD; *P < .05.
We wondered whether cotreatment of the prodifferentiation agents ATO or ATRA with 1 of the pharmacological screening-derived compounds might cooperatively increase CD20 expression as the basis for rituximab or tositumomab activity. Therefore, we exposed L1236 cells, in which ATO strongly enhanced CD20 and compound 40 markedly induced CD19 expression (compare Figures 2B, 4C-D), to very low doses of either ATRA (20 μM) or ATO (5 μM) alone or in combination with compound 40. Strikingly, the already robustly ATRA- or ATO-induced CD20 transcript levels almost doubled, when the cells were coexposed to compound 40 (Figure 5D). This effect was reproducible on the protein level by immunoblot analysis as well. Moreover, CD20 membrane expression became detectable by immunofluorescence image-based flow cytometry upon combined ATO/40 treatment, although the signal was less intense and of a somewhat more aggregated pattern when compared with the B-NHL cell line SU-DHL4 (Figure 5E). Conventional flow cytometry confirmed the increased signal intensity in ATO/40-preexposed as compared with untreated L1236 cells (Figure 5F). Hence, we decided to test whether the most effective combination, ATO plus compound 40, enhancing CD20 transcript expression by nearly 20-fold and producing a positive CD20 signal at the cell surface, might further promote rituximab- or tositumomab-induced cell death. Notably, ATO was used here at the low concentration of 5 µM to reduce potential toxicity of the triple-agent regimen. Indeed, when ATO was administered in combination with compound 40 prior to addition of the CD20 antibody, both rituximab or tositumomab exerted a much stronger cell-autonomous cytotoxic effect as compared with ATO-only– or compound 40-only–preexposed and antibody-treated L1236 cells (Figure 5G; see supplemental Figure 6A for similar results obtained with L428 cells). Given the almost doubled efficacy of rituximab- or tositumomab-induced cell death by the addition of compound 40 plus ATO in this short-term cytotoxicity in vitro assay, we speculate that the actual in vivo activity of the triple-agent anti-CD20 principle may be even more pronounced because additional non-cell-autonomous modes of lymphoma cell death, as demonstrated by a >80% increase in specific cell death upon ATO/compound 40 pretreatment in rituximab-mediated ADCC for L1236 cells and a >190% increase for L428 cells in vitro (Figure 5H; see supplemental Figure 6B for similar results obtained with tositumomab in these cHL cell lines), and immune-mediated clearance of antibody-induced senescent lymphoma cells41,42 further add to the full realization of the antibody-dependent therapeutic benefits in patients.
ATO and compound 40 sensitize cHL cell lines to BCR signaling-targeting kinase inhibitors
The 2 novel kinase inhibitors ibrutinib, targeting the Bruton tyrosine kinase (BTK) in proximal BCR signaling, and idelalisib, blocking the δ-isoform of the also BCR-enhanced phosphatidylinositol 3-kinase (PI3K-P110δ) activity, play increasing roles in the clinical treatment of a variety of B-NHL entities.43-48 Given the marked effects the restoration of B-cell–specific differentiation markers had on CD20-mediated cell death in cHL cells, we sought to explore whether ATO, compound 40, or their combination might also sensitize cHL cells to small compounds that interfere with BCR signaling. Strikingly, L1236 cells that were preexposed to ATO and compound 40 displayed a dramatic susceptibility to ibrutinib-induced cell death that was about 2 orders of magnitude higher when compared with ibrutinib action in otherwise naive L1236 cells (Figure 6A). Notably, sensitization to the BTK inhibitor was also seen after single-agent ATO or compound 40 pretreatment, albeit to a lesser extent, and L428 cells (without exogenous PAX5 restoration) recapitulated this pattern (Figure 6A). Likewise, L1236 cells preincubated with the ATO/40 combination gained extreme sensitivity to idelalisib, exceeding the already enhanced cytotoxic effects the PI3K inhibitor produced in single-agent pretreated L1236 cells (Figure 6B). L428 cells also exhibited the lowest viability in response to the ATO/40-idelalisib sequence. Notably, single-agent ibrutinib or idelalisib has no cell-autonomous efficacy in Hodgkin lymphoma because it produced virtually no toxicity in otherwise untreated cHL cells (viability ≥95%; supplemental Figure 7). To elucidate the underlying molecular mechanism, we probed lysates from L1236 cells that were either untreated or exposed to ATO, compound 40, or both (regarding BTK and PI3K activation, indicated by the phospho-BTK-Tyr223 [p-BTK] and the PI3K downstream target p-AKT-P-Ser473 [p-AKT]) by immunoblot analysis. Indeed, ATO, compound 40, or the combination of both agents had a significant impact on BCR signaling: both the p-BTK-to-total BTK ratio as well as the p-AKT-to-total AKT ratio were strongly induced (Figure 6C; see similar immunoblot findings for L428 cells in supplemental Figure 8). Finally, we sought to test additional cHL cell lines, namely L540, L591, HDML-2, SUP-HD1, and U-HO1, regarding their phenotypic B-cell restorability by the agents used in this study so far: indeed, all compounds induced CD19 and CD20 transcript levels to varying extents in at least some of these cHL cell lines (supplemental Figure 9), thereby underscoring the general applicability of the restoration strategy to the majority of Hodgkin lymphomas tested, while also indicating a certain degree of heterogeneity with respect to the responsiveness of cHL cell lines and very little additional inducibility in B-NHL cell lines when judged by CD19 or CD20 reexpression. Strikingly, the ATO/40 combination rendered all of these additional cHL cell lines dramatically sensitive to the BCR-related pathway blockers (Figure 6D). In essence, our pharmacological hunt for B-cell phenotype-restoring agents not only retrieved working candidates, but further led us to the clinically highly relevant observation that such a pretreatment can be therapeutically exploited with a variety of B-cell–specific antibodies or BCR-signaling inhibitors.
Cytotoxicity of single-agent and combination treatments targeting the BCR-signaling pathway. (A) Relative viability of L1236 (left) and L428 (right) cells exposed to ibrutinib subsequent to the indicated pretreatments (as in Figure 5G). Percentages reflect normalization to control (untreated [UT]; no pretreatment). Ibrutinib was administered at 2.5 nM for 24 hours. Data are presented as mean ± SD; *P < .05. (B) As in panel A, but exposed to idelalisib at 5 μM for 24 hours. (C) Immunoblot analysis to detect p-BTK/BTK and p-AKT/AKT levels in L1236 cells after single-agent or combination treatments or left UT as indicated; α-tubulin as a loading control (left). Relative densitometric values of the scanned immunoblot bands for the p-BTK/BTK (right, top) and p-AKT/AKT ratios (right, bottom), calculated after normalization to the corresponding α-tubulin signal intensities. (D) Heatmap-encoded effects of ibrutinib (left) and idelalisib (right) on the viability of the 8 cHL cell lines after the indicated pretreatments as in panels A and B (color coding for relative viability).
Cytotoxicity of single-agent and combination treatments targeting the BCR-signaling pathway. (A) Relative viability of L1236 (left) and L428 (right) cells exposed to ibrutinib subsequent to the indicated pretreatments (as in Figure 5G). Percentages reflect normalization to control (untreated [UT]; no pretreatment). Ibrutinib was administered at 2.5 nM for 24 hours. Data are presented as mean ± SD; *P < .05. (B) As in panel A, but exposed to idelalisib at 5 μM for 24 hours. (C) Immunoblot analysis to detect p-BTK/BTK and p-AKT/AKT levels in L1236 cells after single-agent or combination treatments or left UT as indicated; α-tubulin as a loading control (left). Relative densitometric values of the scanned immunoblot bands for the p-BTK/BTK (right, top) and p-AKT/AKT ratios (right, bottom), calculated after normalization to the corresponding α-tubulin signal intensities. (D) Heatmap-encoded effects of ibrutinib (left) and idelalisib (right) on the viability of the 8 cHL cell lines after the indicated pretreatments as in panels A and B (color coding for relative viability).
Discussion
Previous studies revealed that epigenetically silenced, transcriptionally repressed, or inadequately induced promoters of B-cell–specific genes may contribute to cHL phenotypic transformation.8,10,12,13,15 We present now, based on a high-throughput screening and further characterization, a number of compounds which share the capacity to restore components of the B-cell phenotype that are consistently lost in cHL cells. Specifically, our data suggest that 2 of these agents exert their effects via remodeling of repressive chromatin marks in the vicinity of the CD19 promoter. The differentiation-enforcing agents ATO and ATRA complement the effects of the screen-identified drugs at a variety of B-cell–specific gene loci, although the precise mechanism of action needs to be elucidated in future investigations and may vary in individual cases. For instance, different expression levels of preferentially antigen expressed in melanoma (PRAME), a repressor of retinoic acid signaling, seem to affect ATRA sensitivity in cHL cell lines.38 Our own findings obtained after derepressing treatment with the DNA-demethylating agent 5-Aza and the histone deacetylase inhibitor TSA support the idea that a master repressor of the B-cell program is selectively active in Hodgkin lymphoma, but epigenetically silenced in B-NHL and normal B cells. We may also have uncovered a putative role of the B-cell phenotype as a tumor-suppressive principle and novel vulnerability of cHL cells: different agents that led to the reexpression of B-cell–specific genes despite their pharmacologically distinct modes of action turned out to be toxic for cHL cells.
The putative targets of the screen-identified compounds point to an important role of epigenetic dysregulation at histone H3 in cHL.32 Both compounds 40 and 49 reportedly inhibit, among other epigenetic regulators, the histone-lysine N-methyltransferase EHMT2 (aka, G9a or KMT1C), which confers transcriptional repression through methylation of H3K9 and less pronouncedly of H3K27.32,34 Indeed, knockdown of EHMT2, comparable to its pharmacological inhibition by BIX-01294, also reinduced CD19 transcript expression in cHL cells (supplemental Figure 10). Although other modes of pharmacological action, indirect and via different or even a variety of target molecules, may apply, it is an attractive hypothesis that the epigenetic changes at the CD19 promoter observed in response to compound 40 or 49 are due to direct inhibition of a histone methylation-modifying enzyme operating at this site.
Most importantly, our findings demonstrate the therapeutic potential restoration of the B-cell program in Hodgkin lymphoma may have (with respect to the increasing arsenal of B-cell–preferential or –exclusive therapeutic options available, and, so far, nonapplicable) to patients diagnosed with cHL. Monoclonal antibodies or antibody-drug conjugates raised against B-cell–specific surface receptors such as CD19, CD20, or CD79 play no role in current (chemo-)immunotherapy regimens used against cHL. Especially in light of the success story of CD20 antibodies in the clinical care of B-NHL, it is a very attractive goal to “repurpose” these antibodies for Hodgkin lymphoma, after restoration of the lost B-cell phenotype by ATO/ATRA and/or compounds identified in our screen. Notably, our data indicate that the derepressed surface expression levels of the CD20 target we achieved on Hodgkin cells, although lower than endogenous levels detectable on B-NHL cells by orders of magnitude, are sufficient to enable CD20 antibody-mediated cytotoxicity. Although feasibility and actual efficacy must be demonstrated in clinical trials, we also like to speculate that the agents discussed here might exert additional benefits in cHL cells beyond restoration of the B-cell program. Although it did not escape our attention that preliminary preclinical and clinical evidence has been reported regarding the PI3K and the BTK inhibitor in the context of Hodgkin lymphoma,49,50 we present here a hitherto unknown strategy to dramatically booster cHL’s susceptibility to BCR-related pathway blockers. Future preclinical investigations will aim at a systematic, multidimensional survey of various combinatorial and sequential treatments (including additional B-cell–targeting antibodies and other compounds) at different concentrations (to also address synergism) in an expanded panel of available cHL cell lines, thereby addressing a larger array of B-cell phenotype-related gene products, and approaching the underlying molecular mechanisms of pharmacological action. Of note, screening-derived lead compounds need to undergo structural optimization by medicinal chemists to lower required doses and potential toxicities before moving toward early-phase clinical testing in relapsed or refractory cHL patients. Taken together, various combinations of B-cell–reprogramming agents, particularly in combination with B-NHL-established antibody- and/or signaling inhibitor-based therapeutics, could open a conceptually novel perspective in clinical care of Hodgkin lymphoma, especially in high-risk or relapsed patients.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Julia Schneider, Carola Seyffarth, and Anja Wolf for technical assistance, Cécile Tonnelle for the pTRIPΔU3-CD19-GFP plasmid, and Mikael Sigvardsson for the mb-1 and B29 promoter reporter construct.
This work was conducted within the Transegional Collaborative Research Center SFB/TRR 54 and was supported by the Deutsche Forschungsgemeinschaft (F.R., S.M., M.H., B.D., S.L., C.A.S.), the German Cancer Aid (Deutsche Krebshilfe Grant no. 110678) to C.A.S., and the German Cancer Consortium (DKTK section “Exploiting Treatment Resistance in Lymphoma” to C.A.S.), and further supported by Helmholtz–China Scholarship Council stipends (J.D., Y.Y.).
Authorship
Contribution: J.D., S.L., J.H.M.D., M.M., G.B., S.R., M.R., Y.Y., N.-R.N., A.B., and P.L. performed experiments; J.D., M.N., N.-R.N., K.S., P.L., E.S., and S.L. analyzed data and compiled the figures; F.R., S.M., M.H., B.D., J.P.v.K., S.L., and C.A.S. designed the research; and C.A.S. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Clemens A. Schmitt, Medical Department of Hematology, Oncology and Tumor Immunology, Charité - Universitätsmedizin Berlin, Augustenburger Platz 1, 13353 Berlin, Germany; e-mail: clemens.schmitt@charite.de.

![Figure 1. Modulation of CD19 promoter activity in cHL and B-NHL cell lines. (A) Expression of the B-cell–specific surface marker CD20 on B-NHL samples (follicular lymphoma [FL], left; diffuse large B-cell lymphoma [DLBCL], right), but not on cHL samples (with CD30 as a typical cHL marker). Standard hematoxylin and eosin (H.E.) staining to visualize morphology. Note CD20− HRS (arrowheads) surrounded by a few infiltrating, CD20+ nonmalignant B cells in the cHL sections. (B) Endogenous CD19 surface antigen expression levels detected by flow cytometry. MFI of individual cHL and B-NHL cell lines (left), and group average of each entity (right). (C) Firefly luciferase-detected CD19 promoter reporter activity, normalized by total protein content in a similar panel of individual (left) and entity-grouped (right) cHL and B-NHL cell lines as in panel B. (D) MFI of CD19 surface antigen expression in cHL and B-NHL cell lines upon 5-Aza/TSA treatment or left untreated (UT) by flow cytometry (individual cell lines, left; average of B-NHL cell lines, right). (E) Firefly luciferase–indicated CD19 promoter activity in cHL and B-NHL cell lines upon 5-Aza/TSA treatment (as in panel D). Data are presented as mean ± SEM. All experiments were done at least in triplicate; *P < .05 throughout the figure.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/129/1/10.1182_blood-2016-02-700773/4/m_blood700773f1.jpeg?Expires=1769118827&Signature=b-Drhfmhmw5L1zgFBMVP1iMQDQBc3AemdmG1v5TpPuBCBq~7w0WT9M-siIFDLtuAWQ0ZaeWv2Y~hbuyQQMq6wC9HeiU1NX8FdvTfvwZdYKHhD5lcOw4b1dgDgUe9ex~nUJjCqPMd7UuO0WdMBp2KoHyvCzbdw6TlkxHBFjnYYf00vqLNXwwteLkcSxCxHhOgibYcjrzcE885KM6Qwi04AillGP9~PZxV9QgU~W~88g2Eu902YOwo~CPleRir87ddRSGvNyAc5j2KN19XmFKvD1RaEyB1N6zVUnXwEfBv508iF0E2xQGOiFCZAi2Jz8DXBC-v~UdSLpLUAc0~t0W1WQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


![Figure 5. Anti-CD20 cytotoxicity after single-agent or combination pretreatment. (A) Relative viability of L1236 cells after exposure to ATO at the indicated concentrations for 48 hours, normalized to the untreated control (UT). (B) As in panel A, but after ATRA. (C) Relative viability of L1236 cells pretreated with 10 µM ATO, 40 µM ATRA, or dimethyl sulfoxide (UT), subsequently exposed to CD20 antibodies rituximab (20 µg/mL) or tositumomab (10 µg/mL) or no antibody for 3 days. Percentages reflect normalization to the no-treatment setting. (D) CD20 expression after single-agent and combination treatments (as indicated) in L1236 cells, at the transcript level (top; relative values by RQ-PCR, normalized to the untreated control [UT]), and at the protein level (bottom; by immunoblot analysis, with α-tubulin as a loading control). ATRA, 20 μM; ATO, 5 μM; compound 40, 10 μM. (E) CD20 surface expression by immunofluorescence image-based flow cytometry (bottom) and matching brightfield pictures (top) in untreated (UT) SU-DHL4 B-NHL cells and L1236 cells exposed to ATO/compound 40 as in panel D (ATO/40), or untreated (UT); shown are 2 representative examples each. Of note, the L1236 signals are displayed with a fourfold amplification compared with the SU-DHL4 signals. (F) CD20 surface expression by conventional flow cytometry in L1236 cells as in panel E. (G) Relative viability of L1236 cells exposed to the CD20 antibodies rituximab or tositumomab (as in panel C) after the indicated pretreatments (as in panel D, ie, ATO at only 5 µM). (H) Rituximab-mediated ADCC values in L1236 (left) or L428 cHL cells (right) preexposed to the indicated drugs or combinations (as in panel G), 6 hours after the addition of the NFAT-luciferase–engineered effector T cells, relative to no preexposure (UT). Data are presented as mean ± SD; *P < .05.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/129/1/10.1182_blood-2016-02-700773/4/m_blood700773f5.jpeg?Expires=1769118827&Signature=gEkxoLs3CCNeFDKzfAD~GIW8u2mpmBQew07qYJMARE7NNQ9oYffNArbFefvahL29ffTru6ixRTkKJOeDrzJ6o9LNIybXJPEF973dxK51zeoAZ2p0jE90vat9jWzrkV7UJd2ztimFNT-WyPNZ9Zw2e1OxTvwl93zcdzCTzTwOpHwfxebYhTkU6B-E-zKzdePM07il0E~YDu1lY3FeDvx9ak1Sbr7XJ7ig1deZQj8JyOHbBdVIvbqXqjMVjWOTfpqEAvb887wW5mFYmbxoIP17lu6hnuBpy70HYZmTrqPwSAcLAmSbMbxq1zHqvazjd~cneRdXuNrlqlrLkJZZ0c4~sw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 6. Cytotoxicity of single-agent and combination treatments targeting the BCR-signaling pathway. (A) Relative viability of L1236 (left) and L428 (right) cells exposed to ibrutinib subsequent to the indicated pretreatments (as in Figure 5G). Percentages reflect normalization to control (untreated [UT]; no pretreatment). Ibrutinib was administered at 2.5 nM for 24 hours. Data are presented as mean ± SD; *P < .05. (B) As in panel A, but exposed to idelalisib at 5 μM for 24 hours. (C) Immunoblot analysis to detect p-BTK/BTK and p-AKT/AKT levels in L1236 cells after single-agent or combination treatments or left UT as indicated; α-tubulin as a loading control (left). Relative densitometric values of the scanned immunoblot bands for the p-BTK/BTK (right, top) and p-AKT/AKT ratios (right, bottom), calculated after normalization to the corresponding α-tubulin signal intensities. (D) Heatmap-encoded effects of ibrutinib (left) and idelalisib (right) on the viability of the 8 cHL cell lines after the indicated pretreatments as in panels A and B (color coding for relative viability).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/129/1/10.1182_blood-2016-02-700773/4/m_blood700773f6.jpeg?Expires=1769118827&Signature=iSVfG3rodqSLWpTXK7akoTQvkFG2I6uf487rW6vAtMDRAniHYA6~kzYefj7oSKWGOsgR-1mN9RfInS~-zlGxTvx6rCuhyn~4AIr-zHbx~ShpajGFoMm7q4jXqHNiomIKuRQU5CmS~Xx~C5o3kiFyAMKruEbn6mGn5waASe-vZKf1hzF2eoIML44I7vvV5fwE9wpg3cXErW-QQIgXsTVYiuNh4pm~ltl9WyA0oKAxHqlugyc-GSuTDaAOZrstHqd9N4ympvZJJVbNObIho6RKEpiZiiaGO3noSDjUvGXNq-20Y1vWhwGYrzcnXsLkPG52~2pkSZcKFLQBc7djBNG85Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)