Key Points
Inactivation of Sin3B in the hematopoietic compartment impairs HSC functions.
Sin3B regulates HSC differentiation and quiescence.
Abstract
Hematopoietic stem cells (HSCs) reside at the top of the hematopoietic hierarchy and are the origin of all blood cells produced throughout an individual’s life. The balance between HSC self-renewal and differentiation is maintained by various intrinsic and extrinsic mechanisms. Among these, the molecular pathways that restrict cell cycle progression are critical to the maintenance of functional HSCs. Alterations in the regulation of cell cycle progression in HSCs invariably lead to the development of hematologic malignancies or bone marrow failure syndromes. Here we report that hematopoietic-specific genetic inactivation of Sin3B, an essential component of the mammalian Sin3–histone deacetylase corepressor complex, severely impairs the competitive repopulation capacity of HSCs. Sin3B-deleted HSCs accumulate and fail to properly differentiate following transplantation. Moreover, Sin3B inactivation impairs HSC quiescence and sensitizes mice to myelosuppressive therapy. Together, these results identify Sin3B as a novel and critical regulator of HSC functions.
Introduction
The ability of hematopoietic stem cells (HSCs) to repopulate the hematopoietic compartment throughout an organism’s life is maintained by discrete intrinsic and extrinsic mechanisms.1,2 Among those, the molecular pathways that restrict HSC cycling and maintain HSC quiescence are critical to the lifelong function of HSCs. In the mouse, lineage-tracing experiments have estimated that each HSC divides only once every 25 to 145 days.3,4 Competitive transplantation studies have demonstrated that quiescent HSCs, but not cycling HSCs, can reconstitute the entire hematopoietic system in vivo.3,5,6 Similarly, in settings of physiological stress such as bleeding or infection, quiescent HSCs are required for the replenishment of the hematopoietic compartment.7 Consistently, gene expression analyses have revealed that proproliferative genes are transcriptionally repressed in HSCs.8 Although the molecular factors that mediate this repression are critical for HSC functions, their identity remains largely unknown.
Previous studies have identified the chromatin-associated Sin3B protein as an important regulator of cell cycle withdrawal in various cellular contexts.9-11 Sin3B is a noncatalytic scaffold protein that serves as a core component for various histone deacetylase (HDAC) transcriptional repressor complexes, which are recruited to genomic loci via the interaction with sequence-specific transcription factors.11,12 We have also recently demonstrated that a Sin3B-containing complex regulates postinitiation transcriptional events, through its interaction with chromatin readers.13 Importantly, genetic manipulation in the mouse revealed that Sin3B and the closely related Sin3A protein are not functionally redundant.14,15 Although Sin3A is required for cellular viability and early embryonic development, Sin3B null embryos survive until late gestation. However, terminal differentiation of specific lineages is impaired upon Sin3B inactivation, resulting in late-embryonic lethality of Sin3B−/− mice.15 In quiescent fibroblasts and hepatocytes, Sin3B is tethered to E2F target loci and represses transcription in a HDAC-dependent manner.15-17 Consistent with these biochemical properties, Sin3B is required for quiescence upon serum starvation and for cellular senescence upon oncogene activation or serial passaging in mouse embryonic fibroblasts.15,18,19 These experiments indicate that Sin3B modulates cell cycle withdrawal and differentiation in somatic cells. However, whether Sin3B controls cell cycle withdrawal in stem cells has not been investigated. Moreover, high Sin3B expression correlates with poor survival in acute myeloid leukemia patients suggesting a role for Sin3B in hematopoietic differentiation and/or proliferation.20
Here, we characterize the contribution of Sin3B to HSC functions. We demonstrate that Sin3B is critical for the competitive repopulation capacity of HSCs, correlating with its role in promoting HSC differentiation. Furthermore, we establish that Sin3B promotes HSC quiescence and protects animals from hematopoietic injury. Together, our study indicates that the corepressor Sin3B engages a transcriptional program that regulates functions central to HSC biology.
Materials and methods
Flow cytometric analysis and cell sorting
Single cell suspensions were derived from bone marrow (femur and tibia of both hind legs), spleen, thymus, and peripheral blood and red blood cells were lysed with ammonium-chloride-potassium lysis buffer. Cell counts were determined using a cell counter (Beckman Coulter) set to detect nuclei between 3.5 μM and 10 μM. All cells were blocked with purified rat immunoglobulin G (IgG; 20 μg/mL) for 15 minutes on ice. The following antibodies were used for analysis (from Biolegend or BD Pharmingen): anti-B220 (RA3-6B2), anti-CD19 (6D5), anti-CD3e (145-2C11), anti-CD4 (RM4-5), anti-CD8 (53-6.7), anti-CD11b (M1-70), anti-Gr1 (RB6-8C5), anti-TER119 (TER119), anti-CD25 (PC61), anti-IgM (RMM-1), anti-IgD (11-26c.2a), anti-CD45.1 (A20), anti-CD45.2 (104), anti-CD16/32 (93), anti-IL7Ra (A7R34), anti-ckit (2B8), anti-Sca1 (D7), anti-Flk2 (A2F10), anti-CD34 (RAM34), and anti-CD150 (TC15-12F12.2). Cells were stained with primary and secondary antibodies for 30 minutes on ice unless the cocktail included anti-CD34 where cells were stained for 90 minutes on ice. For sorting long-term HSCs (LT-HSCs), lineage positive cells were depleted with streptavidin coated dynabeads (Invitrogen) according to the manufacturer’s instructions prior to staining. Propidium iodide (1 μg/mL), 4′,6-diamidino-2-phenylindole (DAPI; 500 ng/mL), or 7-aminoactinomycin D (1 μg/mL) was added following staining for the exclusion of dead cells. Cells were analyzed on a LSRII (BD) or sorted on an AriaI or AriaII (BD). Data were analyzed with FlowJo software (Treestar).
Serial competitive repopulation assay
Lethally irradiated CD45.1 recipient mice (split doses of 6 Gy separated by 4 hours; total body irradiation, cesium source) were injected retroorbitally with a 1:1 mixture of 1 × 106 wild-type CD45.1 whole bone marrow competitor cells and either 1 × 106 wild-type or Sin3BF/F vav1-cre+ CD45.2 whole bone marrow test cells. After 20 weeks, 2 × 106 whole bone marrow cells from primary recipient mice were transplanted into lethally irradiated CD45.1 secondary recipient mice. After 20 weeks, 2 × 106 whole bone marrow cells from secondary recipient mice were transplanted into lethally irradiated CD45.1 tertiary recipient mice. All transplant experiments were performed at least 2 times. Irradiated mice had water supplemented with antibiotics for 4 weeks.
RNA sequencing
RNA was isolated from fluorescence-activated cell sorting (FACS) purified LT-HSCs pooled from 4 animals. RNA was harvested from 2 biological replicates using the RNeasy microkit (Qiagen). RNA quantification and quality were determined using an Aglient 1200 Bioanalyzer. Strand-specific libraries were prepared using the SMARTer low-input RNA kit. Libraries were sequenced on an Illumnia HiSeq2500 using 50 bp paired-end reads. Fastq files were aligned to mm9 using TopHat. Differential expression tests were done using the Cuffdiff module of Cufflinks against the Refseq annotation. We used an false discovery rate (FDR) <0.1 as a cutoff for significance. Gene set enrichment analysis (GSEA) was performed using log2 fold change as metric for ranking genes and 1000 permutations. Gene sets used in this study were identified from the Molecular Signatures Database (Curated v5.0 and Hallmarks v5.0). Raw data were deposited in the Gene Expression Omnibus database (accession number GSE83218).
Statistics
Samples were compared by unpaired 2-tailed Student t test. Survival curves were compared with a Mantel-Cox log-rank test.
Results
Hematopoietic deletion of Sin3B promotes the accumulation of HSPCs
Examination of expression data using the Gene Expression Commons database21 in hematopoietic stem and progenitor cells (HSPCs) indicates that Sin3B expression is highest in LT-HSCs and downregulated as cells differentiate into committed progenitor cells (Figure 1A), suggesting that Sin3B may regulate HSC functions. Thus, to elucidate the contribution of Sin3B to HSC biology, we generated mice where Sin3B is genetically inactivated early in development within hematopoietic cells and endothelial cells, using the vav1-Cre transgene.22 For simplicity, Sin3BF/F vav1-cre+ mice are thereafter referred to as Sin3BCKO (Sin3B conditional knockout), and their Sin3BF/F littermates are referred to as control. Analysis of genomic DNA and protein isolated from whole bone marrow demonstrated that Sin3B was efficiently deleted in Sin3BCKO animals (Figure 1B; supplemental Figure 1A, available on the Blood Web site). Animals of all genotypes were born at expected frequencies (data not shown). There was no difference in the cellularity of the bone marrow, spleen, and thymus between adult Sin3BCKO animals (8-12 weeks of age) and their control littermates (Figure 1C). We observed modest differences in the frequencies of B and T cells in the peripheral blood and the spleen of Sin3BCKO mice compared with control animals (Figure 1D-E). Analysis of progenitor T cells revealed a decrease in the frequency of double-positive T cells and an increase in the frequency of CD4 single-positive T cells in Sin3BCKO animals (supplemental Figure 1B). There was no difference in the frequency of progenitor B cells in the bone marrow of control and Sin3BCKO animals (supplemental Figure 1C). When we analyzed HSPC populations in Sin3BCKO animals, we observed a modest but significant increase in the frequency and number of multipotent progenitors, short-term HSCs (ST-HSCs), and LT-HSCs compared with controls (Figure 1F-G). This was not the case for the myeloerythoid restricted progenitor populations or common lymphoid progenitors (Figure 1F-G; supplemental Figure 1D-E), suggesting a more prominent role for Sin3B in HSPCs compared with committed progenitors. It is unlikely that the accumulation of HSPCs in Sin3BCKO animals reflected differences in cell death, as we did not detect any difference in the frequency of apoptotic LSK cells between Sin3BCKO and control animals (supplemental Figure 1F).
HSPCs accumulate in Sin3BCKOmice. (A) Expression pattern of Sin3B obtained from Gene Expression Commons using the Transcriptome Analysis Identifies Regulators of Hematopoietic Stem and Progenitor Cells Model. Numbers indicate the percentage “activity level” based on the dynamic range of microarray probes as described in Seita et al.21 (B) Polymerase chain reaction analysis of genomic DNA isolated from bone marrow mononuclear cells of indicated genotypes. (C) Quantification of the number of cells present in the indicated organs of control and Sin3BCKO animals; n = 6. (D) Frequency of indicated cell populations in the peripheral blood of animals; n = 6. (E) Frequency of indicated cell populations in the spleen of animals; n = 6. (F) Representative FACS plot for various HSPC populations. Mean frequency of indicated cell populations are indicated; n = 6. P < .05 for lin- sca1+ ckit+ (LSK), ST-HSC, and LT-HSC. (G) Quantification of the cell number of indicated HSPC populations; n = 6. Data are represented as mean ± standard error of the mean (SEM). *P < .05; **P < .01. BM, bone marrow; CLP, common lymphoid progenitor; CMP, common myeloid progenitor; GMP, granulocyte-macrophage progenitor; LP, lineage positive; MEP, megakaryocyte-erythroid progenitor; MP, myeloerythoid progenitor; MPP, multipotent progenitor.
HSPCs accumulate in Sin3BCKOmice. (A) Expression pattern of Sin3B obtained from Gene Expression Commons using the Transcriptome Analysis Identifies Regulators of Hematopoietic Stem and Progenitor Cells Model. Numbers indicate the percentage “activity level” based on the dynamic range of microarray probes as described in Seita et al.21 (B) Polymerase chain reaction analysis of genomic DNA isolated from bone marrow mononuclear cells of indicated genotypes. (C) Quantification of the number of cells present in the indicated organs of control and Sin3BCKO animals; n = 6. (D) Frequency of indicated cell populations in the peripheral blood of animals; n = 6. (E) Frequency of indicated cell populations in the spleen of animals; n = 6. (F) Representative FACS plot for various HSPC populations. Mean frequency of indicated cell populations are indicated; n = 6. P < .05 for lin- sca1+ ckit+ (LSK), ST-HSC, and LT-HSC. (G) Quantification of the cell number of indicated HSPC populations; n = 6. Data are represented as mean ± standard error of the mean (SEM). *P < .05; **P < .01. BM, bone marrow; CLP, common lymphoid progenitor; CMP, common myeloid progenitor; GMP, granulocyte-macrophage progenitor; LP, lineage positive; MEP, megakaryocyte-erythroid progenitor; MP, myeloerythoid progenitor; MPP, multipotent progenitor.
Sin3B is required for the competitive repopulation capacity of HSCs
The previous observations prompted us to investigate whether Sin3B regulates pathways critical for HSC functions. Initially, we sorted LT-HSCs and cultured them in myeloid and erythroid-promoting methylcellulose. There was no difference in the ability of Sin3BCKO or control LT-HSCs to form colonies at D7 and D10 of culture (supplemental Figure 2A-B). Moreover, Sin3BCKO LT-HSCs were not defective at differentiating into specific myeloid or erythoid lineages in culture (supplemental Figure 2C). However, we observed a significant decrease in the ability of Sin3BCKO LT-HSC cultures to form colonies following secondary plating (supplemental Figure 2D) suggesting that Sin3BCKO HSCs are functionally impaired compared with their wild-type counterparts. To formally test this, we performed a competitive repopulation assay by transplanting either control or Sin3BCKO bone marrow (CD45.2) together with an equal amount of wild-type competitor bone marrow (CD45.1) into irradiated recipient animals (CD45.1) (Figure 2A). Although control test cells contributed to roughly 50% of the peripheral blood in recipient animals at 16 weeks posttransplant, Sin3BCKO bone marrow cells were drastically impaired in their ability to contribute to both the lymphoid and the myeloid lineages in the peripheral blood (Figure 2B-E). Similarly, Sin3BCKO-derived cells were found at low frequencies in all hematopoietic organs of primary recipients 20 weeks following transplantation (supplemental Figure 2E). Importantly, Sin3BCKO lineage-depleted bone marrow cells were able to migrate to the bone marrow of recipient animals 16 hours after transplantation at a rate similar to that of control cells, suggesting that the reconstitution defect is unlikely because of Sin3BCKO cells being unable to home to the bone marrow (Figure 2F). Moreover, when we characterized Sin3BCKO-derived cells in the bone marrow of the recipient animals, we detected an accumulation of Sin3BCKO-derived cells within the HSPC compartment and most noticeably within LT-HSCs and ST-HSCs (Figure 2G). Although Sin3BCKO-derived cells represented <10% of the total bone marrow, they accounted for roughly 40% of LT-HSCs and 30% of ST-HSCs (Figure 2H). This characteristic of transplanted Sin3B-deleted hematopoietic cells is consistent with the accumulation of HSPCs we observed in Sin3BCKO animals (Figure 1). Together, these data suggest that although Sin3BCKO HSCs are able to properly home to and engraft in the bone marrow, and to give rise to phenotypic HSCs, they fail to differentiate into committed progenitors and subsequently mature blood types.
Sin3B is required for the competitive repopulation capacity of HSCs. (A) Schematic for serial competitive repopulation assay. Test CD45.2 bone marrow (control or Sin3BCKO) was competed at a 1:1 ratio against wild-type CD45.1 bone marrow and injected intravenously into a lethally irradiated (12 Gy) host. Recipient animals were euthanized after 20 weeks for analysis, and 2 × 106 bone marrow cells were serially transplanted into secondary recipients. Secondary recipients were euthanized after 20 weeks and 2 × 106 bone marrow cells were serially transplanted into tertiary recipients. (B) Quantification of donor-derived (CD45.2) cells in the peripheral blood of recipient animals at indicated time points; n = 8. Quantification of donor-derived (CD45.2) cells of B (B220+) cells (C), T (CD4+ or CD8+) cells (D), and myeloid (CD11b+) cells (E) in the peripheral blood of recipient animals; n = 8. (F) Percent of CD45.2 cells in the bone marrow 16 hours after injection of 2 × 106 lineage depleted bone marrow cells into irradiated CD45.1 mice; n = 5. (G) Representative FACS plot of donor-derived (CD45.2) HSPC populations. (H) Donor contribution of indicated cell populations in the bone marrow 20 weeks after transplantation of primary recipient animals. (I) Donor-derived B (B220+), T (CD4+ or CD8+), and myeloid (CD11b+) cells in the peripheral blood of secondary recipient (left) and tertiary recipient (right) animals; n = 4. (J) Donor contribution of indicated cell populations in the bone marrow 20 weeks after transplantation of tertiary recipient animals; n = 4. Data are represented as mean ± SEM. *P < .05; **P < .01.
Sin3B is required for the competitive repopulation capacity of HSCs. (A) Schematic for serial competitive repopulation assay. Test CD45.2 bone marrow (control or Sin3BCKO) was competed at a 1:1 ratio against wild-type CD45.1 bone marrow and injected intravenously into a lethally irradiated (12 Gy) host. Recipient animals were euthanized after 20 weeks for analysis, and 2 × 106 bone marrow cells were serially transplanted into secondary recipients. Secondary recipients were euthanized after 20 weeks and 2 × 106 bone marrow cells were serially transplanted into tertiary recipients. (B) Quantification of donor-derived (CD45.2) cells in the peripheral blood of recipient animals at indicated time points; n = 8. Quantification of donor-derived (CD45.2) cells of B (B220+) cells (C), T (CD4+ or CD8+) cells (D), and myeloid (CD11b+) cells (E) in the peripheral blood of recipient animals; n = 8. (F) Percent of CD45.2 cells in the bone marrow 16 hours after injection of 2 × 106 lineage depleted bone marrow cells into irradiated CD45.1 mice; n = 5. (G) Representative FACS plot of donor-derived (CD45.2) HSPC populations. (H) Donor contribution of indicated cell populations in the bone marrow 20 weeks after transplantation of primary recipient animals. (I) Donor-derived B (B220+), T (CD4+ or CD8+), and myeloid (CD11b+) cells in the peripheral blood of secondary recipient (left) and tertiary recipient (right) animals; n = 4. (J) Donor contribution of indicated cell populations in the bone marrow 20 weeks after transplantation of tertiary recipient animals; n = 4. Data are represented as mean ± SEM. *P < .05; **P < .01.
Sin3B is required for HSC differentiation following serial transplantation
We next assessed whether the defects of Sin3BCKO-derived LT-HSCs observed after 1 transplant were exacerbated following serial transplantation. To do so, we harvested whole bone marrow from primary recipient animals 20 weeks after the primary transplant and transplanted 2 × 106 bone marrow cells into irradiated secondary recipients. After another 20 weeks, we transplanted 2 × 106 bone marrow cells from secondary recipients into irradiated tertiary recipients (Figure 2A). Similar to our observations following primary transplantation, Sin3BCKO-derived cells contributed only marginally to the peripheral blood in secondary recipients and failed to contribute to the peripheral blood in tertiary recipients, pointing to an exacerbated loss of HSC function over time (Figure 2I). Because of the decreased contribution following tertiary transplantation, it remained possible that Sin3BCKO HSCs exhausted prematurely. However, Sin3BCKO-derived cells contributed to roughly 50% of LT-HSCs, a contribution similar to that seen in controls (Figure 2J). The proportion of cells derived from Sin3BCKO bone marrow cells dropped off drastically in the cellular compartments downstream of LT-HSCs and accounted for <1% of the total bone marrow in tertiary transplanted mice (Figure 2J). Together, these observations indicate that although Sin3BCKO HSCs are capable of deriving phenotypic HSCs over serial transplants, they fail to differentiate into mature blood cells in a competitive transplant setting.
Sin3B null HSCs are myeloid biased
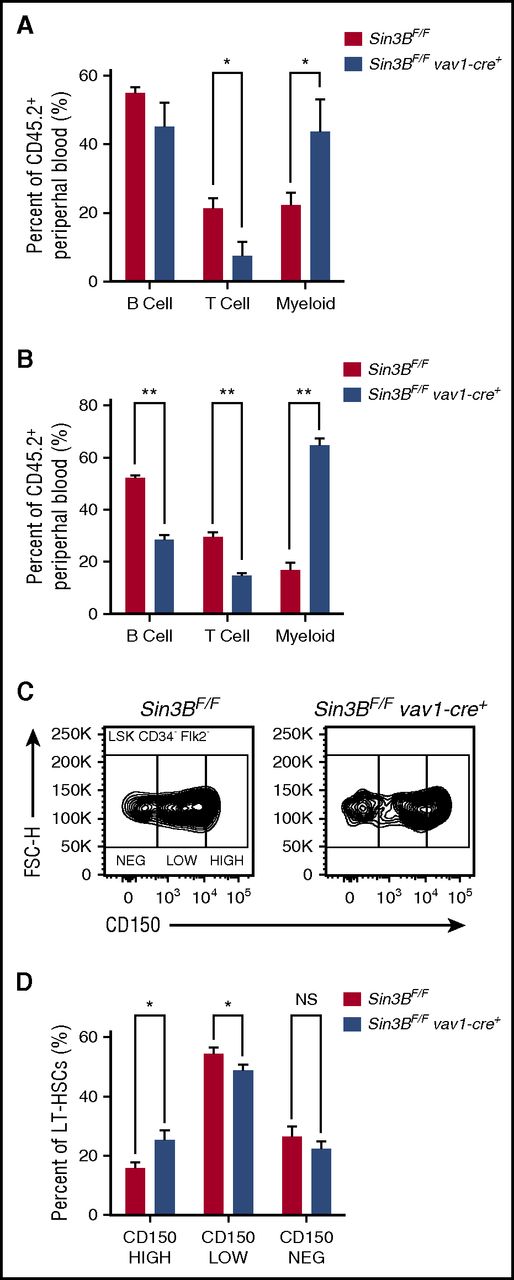
Although only contributing to a minority of differentiated hematopoietic cells following transplantation, Sin3BCKO-transplanted cells preferentially gave rise to mature cells of the myeloid lineages compared with the lymphoid lineages. This bias was detected in the peripheral blood and in the spleen of primary recipient animals (Figure 3A; supplemental Figure 3A) and was further exacerbated following secondary transplantation (Figure 3B). Recent reports have described that within the HSC pool, different types of HSCs give rise to specific lineages.23,24 One type of HSC is the myeloid-biased HSC, which accumulates in aged animals and exhibits higher cell surface expression of CD150 (Slamf1).25,26 As we observed an increase in the ratio of myeloid vs lymphoid cells derived from Sin3BCKO HSCs following transplantation, we assessed the surface expression of CD150 by flow cytometry on Sin3BCKO HSCs prior to transplantation (see gating strategy in supplemental Figure 3B). Consistently, the proportion of CD150HIGH HSCs in the Sin3BCKO HSC compartment was increased compared with controls (Figure 3C-D), suggesting that Sin3B loss correlates with a skewing of the HSC pool toward myeloid-biased HSCs.
Sin3BCKOHSCs are myeloid biased. Percent of B (B220+), T (CD4+ or CD8+), and myeloid (CD11b+) cells from donor-derived peripheral blood in primary (A) and secondary (B) recipient animals 20 weeks after transplantation. (C) Representative FACS plot of CD150 surface staining of LT-HSCs of indicated genotypes. (D) Quantification of percentage of CD150HIGH, CD150LOW, and CD150NEG within the LT-HSC population; n ≥ 8. Data are represented as mean ± SEM. *P < .05; **P < .01. NS, not significant.
Sin3BCKOHSCs are myeloid biased. Percent of B (B220+), T (CD4+ or CD8+), and myeloid (CD11b+) cells from donor-derived peripheral blood in primary (A) and secondary (B) recipient animals 20 weeks after transplantation. (C) Representative FACS plot of CD150 surface staining of LT-HSCs of indicated genotypes. (D) Quantification of percentage of CD150HIGH, CD150LOW, and CD150NEG within the LT-HSC population; n ≥ 8. Data are represented as mean ± SEM. *P < .05; **P < .01. NS, not significant.
Sin3B regulates the transcriptional networks associated with long-term quiescent HSCs
To identify the molecular pathways regulated by Sin3B in HSCs, we performed genome-wide expression profiling on isolated LT-HSCs. Using standard criteria (fold change >1.5; FDR <0.1), we identified 43 genes significantly upregulated and 22 genes significantly downregulated in Sin3BCKO HSCs (Figure 4A). Several of the genes affected by Sin3B loss, including Gata-1, Stat1, and Alcam, have previously been described to play important functions in HSC biology.27-29 Furthermore, and consistent with the phenotypes we observed in vivo, GSEA30 revealed that gene signatures associated with adult stem cells and HSCs negatively correlated with the transcriptional signature of Sin3BCKO HSCs (Figure 4B). We also observed that genes downregulated in quiescent HSCs were enriched in Sin3BCKO HSCs, suggesting that loss of Sin3B results in the derepression of proproliferative genes and entry into cell cycle. In agreement, the Sin3BCKO HSC signature was enriched for genes in the Reactome G1-S transition gene set. Data sets for E2F and Myc targets were also enriched in Sin3BCKO HSCs (Figure 4B). Using chromatin immunoprecipitation enrichment analysis (ChEA),31 the genes significantly dysregulated in Sin3BCKO HSCs were found in previous chromatin immunoprecipitation studies to be bound by transcription regulators previously described as Sin3B interactors, including E2F4 and KDM5A (Figure 4C).32,33 Work in terminally differentiated cells have previously demonstrated that Sin3B associates with E2F4, KDM5A, and HDAC1 at cell cycle genes to repress their transcription.9,16,18 Therefore, we assessed whether Sin3B similarly interacted with these partners in hematopoietic progenitor cells. Because of the low number of LT-HSCs in vivo, we used the HSPC-like HPC-7 cell line. Similar to HSCs, HPC-7 cells rely on stem cell factor for their growth and have multipotent differentiation potential.34 Immunoprecipitation experiments revealed that tagged Sin3B associated with E2F4, KDM5A, and HDAC1 but not with HDAC3 in HPC-7 cells, reminiscent of what has been described in differentiated cells (Figure 4D).9,11,18 Together these data support the notion that Sin3B containing complexes are critical for the gene expression program central to HSC biology.
Sin3B-regulated transcriptional networks in HSCs. (A) Heat map depicting significantly dysregulated genes in Sin3BCKO HSCs compared with control (FDR <0.1; fold change >1.5). (B) GSEA of control and Sin3BCKO HSCs. FWER, family wise error rate; NES, normalized enrichment score. (C) ChEA of significantly dysregulated genes in Sin3BCKO HSCs. (D) FLAG immunoprecipitation (IP) was performed in HPC-7 cells infected with MSCV-IRES-GFP (EV) or MSCV-Sin3B FLAG-IRES-GFP (Sin3B FLAG). Representative immunoblot analysis is shown.
Sin3B-regulated transcriptional networks in HSCs. (A) Heat map depicting significantly dysregulated genes in Sin3BCKO HSCs compared with control (FDR <0.1; fold change >1.5). (B) GSEA of control and Sin3BCKO HSCs. FWER, family wise error rate; NES, normalized enrichment score. (C) ChEA of significantly dysregulated genes in Sin3BCKO HSCs. (D) FLAG immunoprecipitation (IP) was performed in HPC-7 cells infected with MSCV-IRES-GFP (EV) or MSCV-Sin3B FLAG-IRES-GFP (Sin3B FLAG). Representative immunoblot analysis is shown.
Sin3B is required for HSC quiescence
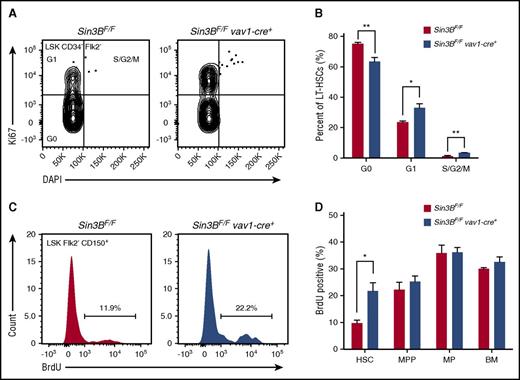
The maintenance of HSC quiescence is critical for HSC functions.5,7 Based on our transcriptome analysis indicating that Sin3BCKO HSCs are enriched for genes associated with cell cycle progression and the demonstration that Sin3B interacts with both HDAC1, E2F4, and KDM5A in HPC-7 cells, we hypothesized that Sin3B regulates HSC quiescence. First, we documented the impact of genetic Sin3B inactivation on the cell cycle status of HSCs using Ki67 and DAPI staining. Sin3BCKO mice exhibited a decreased proportion of quiescent HSCs and, conversely, an increase in the proportion of HSCs in the G1/S/G2/M phases of the cell cycle (Figure 5A-B). Consistently, Sin3BCKO HSCs were more likely to incorporate 5-bromo-2′-deoxyuridine (BrdU) 24 hours following administration than their control counterparts, pointing to an increase in the frequency of actively cycling HSCs (Figure 5C-D). Together, these experiments demonstrate that Sin3B is required for the maintenance of HSC quiescence.
Sin3B is required for the maintenance of HSC quiescence. (A) Representative FACS plot of intracellular Ki67 staining and DNA content (DAPI) in control and Sin3BCKO HSCs. (B) Quantification of the distribution of HSCs in the G0, G1, and S/G2/M phases of the cell cycle; n = 4. (C) Representative FACS plot of BrdU incorporation in control and Sin3BCKO HSCs. (D) Quantification of BrdU incorporation of indicated cell populations; n = 3. Data are represented as mean ± SEM. *P < .05; **P< .01.
Sin3B is required for the maintenance of HSC quiescence. (A) Representative FACS plot of intracellular Ki67 staining and DNA content (DAPI) in control and Sin3BCKO HSCs. (B) Quantification of the distribution of HSCs in the G0, G1, and S/G2/M phases of the cell cycle; n = 4. (C) Representative FACS plot of BrdU incorporation in control and Sin3BCKO HSCs. (D) Quantification of BrdU incorporation of indicated cell populations; n = 3. Data are represented as mean ± SEM. *P < .05; **P< .01.
Sin3B protects mice from 5-FU–mediated myelosuppression
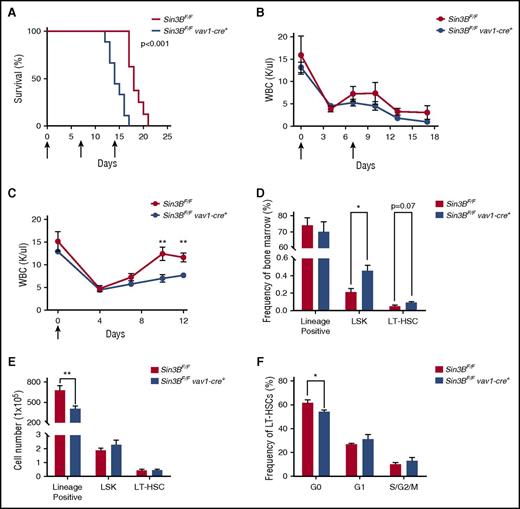
HSCs, because of their ability to differentiate into all hematopoietic lineages and their dormant nature, are essential for the replenishment of the hematopoietic compartment following various stresses such as myelotoxic chemotherapies and infection.7,35 Therefore, we examined whether the impairment of Sin3BCKO HSCs to differentiate as well as their inability to maintain quiescence correlated with increased sensitivity to 5-fluorouracil (5-FU), a myelosuppressive chemotherapeutic. Indeed, following weekly 5-FU injections Sin3BCKO mice died faster than controls (Figure 6A). We confirmed these observations by performing experiments with 2 serial injections of 5-FU and documented the impact of Sin3B inactivation on the circulating blood of these mice. The peripheral blood counts of Sin3BCKO animals were consistently lower than controls, correlating with their decreased survival (Figure 6B; supplemental Figure 4A). To further investigate why Sin3BCKO animals were sensitized to 5-FU treatment, we injected control and Sin3BCKO animals with 1 dose of 5-FU and monitored their recovery. Following 5-FU injection, the peripheral blood counts of Sin3BCKO and control animals dropped to comparable levels. However, we observed a slower recovery of peripheral blood counts in Sin3BCKO animals (Figure 6C). Although there were differences in the frequency of specific cell types in the peripheral blood at different time points following 5-FU injections (similar to what we observed in Sin3BCKO animals at homeostasis; Figure 1D), we did not observe differences in the depletion and/or recovery between specific lineages following both serial and single 5-FU injections. These results suggest that defects in the blood counts of Sin3BCKO mice are not because of impaired differentiation of specific lineages (supplemental Figure 4B-C). Interestingly, we observed an increase in the frequency of LSKs and LT-HSCs in Sin3BCKO animals, but no difference in their cell number compared with controls. However, the number of lineage positive cells in the bone marrow and total splenocytes of Sin3BCKO animals was significantly decreased (Figure 6D-E; supplemental Figure 4D), consistent with a defect in the ability of Sin3BCKO HSPCs to adequately respond and differentiate in order to reestablish homeostasis following stress. Finally, we analyzed the cell cycle profiles of LT-HSCs at day 12 following 5-FU injection and observed a modest decrease in the frequency of quiescent LT-HSCs in Sin3BCKO animals compared with controls (Figure 6F). These data demonstrate a critical role for Sin3B in protecting animals from 5-FU–driven myelosuppression and further confirm the contribution of Sin3B in promoting HSC differentiation and restricting HSC proliferation.
Sin3BCKOanimals are sensitized to myelosuppressive therapy. (A) Survival analysis of control and Sin3BCKO animals injected weekly with 150 mg/kg 5-FU. Arrows indicate days of injection (days 0, 7, and 14); n = 9 per genotype. (B) Absolute leukocyte count in the peripheral blood after serial injections of 5-FU. Arrows indicate days of injection (days 0 and 7). Days 0 and 7 counts are prior to 5-FU injection; n ≥ 3. (C) Absolute leukocyte count in the peripheral blood after a single injection of 5-FU. Arrow indicates day of injection (day 0). Day 0 counts are prior to 5-FU injection; n ≥ 3. Frequency (D) and cell number (E) of indicated populations in the bone marrow at day 12 following 5-FU injection; n ≥ 3. (F) Quantification of the distribution of HSCs in the G0, G1, and S/G2/M phases of the cell cycle at day 12 following 5-FU injection; n ≥ 3. Data are represented as mean ± SEM. *P < .05; **P < .01.
Sin3BCKOanimals are sensitized to myelosuppressive therapy. (A) Survival analysis of control and Sin3BCKO animals injected weekly with 150 mg/kg 5-FU. Arrows indicate days of injection (days 0, 7, and 14); n = 9 per genotype. (B) Absolute leukocyte count in the peripheral blood after serial injections of 5-FU. Arrows indicate days of injection (days 0 and 7). Days 0 and 7 counts are prior to 5-FU injection; n ≥ 3. (C) Absolute leukocyte count in the peripheral blood after a single injection of 5-FU. Arrow indicates day of injection (day 0). Day 0 counts are prior to 5-FU injection; n ≥ 3. Frequency (D) and cell number (E) of indicated populations in the bone marrow at day 12 following 5-FU injection; n ≥ 3. (F) Quantification of the distribution of HSCs in the G0, G1, and S/G2/M phases of the cell cycle at day 12 following 5-FU injection; n ≥ 3. Data are represented as mean ± SEM. *P < .05; **P < .01.
Discussion
The mechanisms that maintain the balance between self-renewal and differentiation in HSCs are critical to prevent the emergence of bone marrow failure syndromes and myeloproliferative diseases. However, the identity of these safeguard mechanisms as well as their regulation remain incompletely understood. Here we identify the corepressor Sin3B as a novel regulator of HSC functions. We demonstrate that loss of Sin3B in the hematopoietic compartment drastically impairs the repopulation capacity of HSCs because of their inability to differentiate into mature blood cells. Furthermore, we found that Sin3B regulates the transcriptional signature associated with quiescent LT-HSCs. Sin3BCKO HSCs are enriched for genes associated with proliferation, and in agreement, Sin3B is required for the maintenance of HSC quiescence. Finally, we observed that hematopoietic loss of Sin3B sensitizes animals to 5-FU–mediated myelosuppression.
Previous studies have demonstrated that chromatin-modifying complexes play important functions in HSCs.36 Work by our laboratory and others have established that Sin3B is an integral component of distinct complexes that mediate discrete biological functions.13,15,17 The prominent Sin3B-containing complex is the canonical Sin3-HDAC complex, which also includes the highly related Sin3A protein. It has been previously shown that inactivation of Sin3A or combined genetic deletion of HDAC1 and HDAC2 results in the drastic loss of hematopoietic cells and loss of HSC viability and repopulation capacity.37 This phenotype differs from what we observed upon genetic inactivation of Sin3B in the hematopoietic system, leading us to speculate that Sin3A and Sin3B regulate distinct pathways in HSCs. In support of this, Sin3A is unable to compensate for the defects present in Sin3BCKO HSCs despite being expressed in these cells (data not shown). It is tempting to speculate that Sin3B associates with specific transcription factors to tether the Sin3-HDAC complex to their target genes, a function that cannot be fulfilled by Sin3A, or alternatively, that an independent Sin3B-containing complex is responsible for the phenotypes observed here. In this regard, the recent characterization of a Sin3B-specific complex, which contains Pf1, EMSY, Mrg15, and KDM5A, but not Sin3A, may rationalize these observations.13,33 This hypothesis is supported by our transcriptome analysis where KDM5A was a top hit following ChEA analysis of Sin3B-regulated genes, our finding that Sin3B associates with KDM5A in HPC-7 cells, and a recent study that identifies KDM5A as a regulator of HSCs.38
Significant work has begun to dissect the molecular underpinnings of Sin3-driven regulation of gene expression and its impacts on cellular processes. Biochemical studies have identified interactions between Sin3-HDAC complexes, E2F4, and the pocket proteins Rb1, p107, and p130.32 Work in cancer cell lines and fibroblasts demonstrated that Sin3B, along with HDAC1, E2F4, and p130, associate at numerous E2F target genes to compact the surrounding chromatin and repress gene expression during quiescence.16,17 Accordingly, loss of Sin3B in both mouse embryonic fibroblasts and hepatocytes results in increased acetylation of E2F target loci and increased expression of these genes, and ultimately prevents cells from exiting cell cycle.15,18 We found that Sin3BCKO HSCs are enriched for transcripts corresponding to proproliferative cell cycle and E2F target genes and fail to properly withdrawal from cell cycle. Moreover, in silico analysis identified E2F4 as a potential candidate for mediating the transcriptional defects of Sin3BCKO HSCs. Consistent with this possibility, Sin3B associates with both E2F4 and HDAC1 in HPC-7 cells. This is further supported by the observation that loss of E2F4, as well as the pocket proteins, impairs the competitive repopulation capacity of HSCs.39-41
Work by van Oevelen et al10,42 revealed that both Sin3A and Sin3B function with E2F4 and KDM5A to permanently repress cell cycle genes in terminally differentiated myotubes and that Sin3 complexes are critical for muscle differentiation in the mouse. Similarly, a recent report described coordinated repression between E2F4 and KDM5A of cell cycle genes in mouse embryonic stem cells as they differentiate.43 Our findings that Sin3B is critical for HSC cell cycle withdrawal and that Sin3B associates with both E2F4 and KDM5A in HPC-7 cells supports the hypothesis that a similar complex containing Sin3B, E2F4, and KDM5A may be critical for HSC differentiation and quiescence. It will therefore be important to systematically identify Sin3B-containing complexes and use biochemical and genetic approaches to dissect their specific contributions to HSC functions.
Our results suggest that loss of Sin3B affects stress hematopoiesis more profoundly than steady-state hematopoiesis similar to observations made for E2F4 and Rb-1.39,40 This is consistent with our studies in fibroblasts demonstrating a more prominent role for Sin3B following stress (including oncogenic stress and replicative stress) as opposed to normal culture conditions.15,18 Furthermore, several lineage-tracing studies have suggested that HSCs are not major contributors to steady-state hematopoiesis, especially in young (8-12 week) animals.35,44-46 This is in contrast to what has been described for stress situations, where HSCs are essential. As such, we hypothesized that aged Sin3BCKO animals may reveal more prominent hematopoietic defects. However, when we analyzed the bone marrow of 1-year-old Sin3BCKO animals, we noted the emergence of wild-type hematopoietic cells (data not shown). Such cells likely derive from HSCs with incomplete vav1-Cre-driven deletion of the Sin3B locus, which then outcompete their Sin3B-deleted counterparts. Together with the demonstration that Sin3BCKO HSCs are impaired in competitive transplantation assays and are myeloid biased, both phenotypes of aged HSCs,47 this observation points to a potential impact of Sin3B loss in an aged setting.
This study complements several other analyses of mice inactivated for cell cycle control genes and identifies Sin3B as a novel regulator of HSCs.48 Although similarities exist between the phenotypes elicited by Sin3B inactivation and those reported for mice with genes in the Rb-E2F pathway deleted (see previous discussion), key differences remain and point to a Sin3B-dependent molecular program independent of the Rb-E2F canonical pathway. It will be of particular interest to further examine whether a single Sin3B-containing complex mediates all the HSC phenotypes we observed or whether numerous complexes regulate distinct aspects. Because chromatin-modifying complexes have been successfully targeted to treat a wide array of diseases,49 a greater understanding of Sin3B-containing complexes and their roles in HSCs may point to novel strategies to improve stem cell transplantation and/or treat hematopoietic diseases.
The data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE83218).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are grateful to all members of the David laboratory for helpful discussions during the preparation of the manuscript and wish to acknowledge the Skirball Institute of Biomolecular Medicine for hosting our laboratory following Hurricane Sandy. The authors thank Michelle Pagano for the generous gift of the E2F4 antibody, the NYU Cytometry and Cell Sorting Core (supported in part by a National Institutes of Health, National Cancer Institute support grant P30CA016087) for help with analysis and cell sorting, and the NYU Genome Technology Center (supported in part by a National Institutes of Health, National Cancer Institute support grant P30CA016087) for help with RNA sequencing and bioinformatic analysis.
This work was supported by the National Institutes of Health, National Cancer Institute (grants R01CA148639 and R21CA155736) (G.D.), the Samuel Waxman Cancer Research Foundation (G.D.), and a Feinberg New York University individual grant (G.D.). D.J.C. was supported by a predoctoral National Institutes of Health, National Cancer Institute training grant (T32CA009161) and a predoctoral National Institutes of Health, National Cancer Institute National Research Service Award (F30CA203047).
Authorship
Contribution: D.J.C. designed the study, performed the experiments, analyzed the data, and cowrote the manuscript; G.D. directed the project and cowrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Gregory David, Department of Biochemistry and Molecular Pharmacology, New York University, MSB417, 550 First Ave, New York, NY 10016; e-mail: gregory.david@nyumc.org.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal