Key Points
An expression map of HSPC differentiation from single-cell RNA sequencing of HSPCs provides insights into blood stem cell differentiation.
A user-friendly Web resource provides access to single-cell gene expression profiles for the wider research community.
Abstract
Maintenance of the blood system requires balanced cell fate decisions by hematopoietic stem and progenitor cells (HSPCs). Because cell fate choices are executed at the individual cell level, new single-cell profiling technologies offer exciting possibilities for mapping the dynamic molecular changes underlying HSPC differentiation. Here, we have used single-cell RNA sequencing to profile more than 1600 single HSPCs, and deep sequencing has enabled detection of an average of 6558 protein-coding genes per cell. Index sorting, in combination with broad sorting gates, allowed us to retrospectively assign cells to 12 commonly sorted HSPC phenotypes while also capturing intermediate cells typically excluded by conventional gating. We further show that independently generated single-cell data sets can be projected onto the single-cell resolution expression map to directly compare data from multiple groups and to build and refine new hypotheses. Reconstruction of differentiation trajectories reveals dynamic expression changes associated with early lymphoid, erythroid, and granulocyte-macrophage differentiation. The latter two trajectories were characterized by common upregulation of cell cycle and oxidative phosphorylation transcriptional programs. By using external spike-in controls, we estimate absolute messenger RNA (mRNA) levels per cell, showing for the first time that despite a general reduction in total mRNA, a subset of genes shows higher expression levels in immature stem cells consistent with active maintenance of the stem-cell state. Finally, we report the development of an intuitive Web interface as a new community resource to permit visualization of gene expression in HSPCs at single-cell resolution for any gene of choice.
Introduction
Hematopoietic stem cells (HSCs) sit at the apex of a differentiation hierarchy that produces the full spectrum of mature blood cells via intermediate progenitor stages. For almost 3 decades, researchers have developed protocols for the prospective isolation of increasingly refined hematopoietic stem and progenitor cell (HSPC) populations, reaching purities of more than 50% for long-term repopulating HSCs.1-5 Although these approaches have provided many significant advances, none of the populations purified to date is composed of a single homogeneous cell type, and the purification protocols necessitate the use of restrictive gates to maximize population purity, thus excluding potential transitional cells located outside these gates.
It has long been recognized that a mechanistic understanding of differentiation processes requires detailed knowledge of the changes in gene expression that accompany and/or drive the progression from one cellular state to the next. Conventional bulk expression profiling of heterogeneous populations captures average expression states that may not be representative of any single cell. Recently developed single-cell profiling techniques are able to resolve population heterogeneity6,7 and profile transitional cells when scaled up to large cell numbers.8 Full flow cytometry phenotypes can be recorded by using index sorting9 to link single-cell gene expression profiles with single-cell function.10 Single-cell profiling also enables reconstruction of regulatory network models11-13 and inference of differentiation trajectories.8,14
Web interfaces that provide access to comprehensive transcriptomic resources have been instrumental in supporting research into the molecular mechanisms of normal and malignant hematopoiesis.15-20 However, there is no comparable resource or Web interface for single HSPC transcriptome data at this time. Here, we present 1656 single HSPC transcriptomes analyzed by single-cell RNA sequencing (scRNA-seq) with broad gates, deep sequencing, and index sorting to retrospectively identify populations by surface marker expression. The resulting single-cell resolution gene expression landscape has been incorporated into a freely accessible online resource that can be used to visualize HSC-to-progenitor transitions, highlight putative lineage branching points, and identify lineage-specific transcriptional programs.
Methods
scRNA-Seq
HSPCs were collected from the bone marrow of 10 female 12-week-old C57BL/6 mice over 2 consecutive days, with cells from 4 mice pooled together and cells from 1 mouse analyzed separately each day. The bone marrow was lineage depleted by using the EasySep Mouse Hematopoietic Progenitor Cell Enrichment Kit (STEMCELL Technologies). The following antibodies were used: anti-EPCR-PE (Clone RMEPCR1560 [#60038PE], STEMCELL Technologies), anti-CD48-PB (Clone HM481 [#103418], BioLegend), anti-Lin-BV510 (#19856, STEMCELL Technologies), anti-CD150-PE/Cy7 (Clone TC15012F12.2 [#115914], BioLegend), anti-CD16/32-Alexa647 (Clone 93 [#101314], BioLegend), anti-CKit-APC/Cy7 (Clone 2B8 [#105856], BioLegend), anti-Flk2-PE/Cy5 (Clone A2F10 [#115914], eBioscience), anti-CD34-FITC (Clone RAM34 [#553733], BD Pharmingen), and 4′,6-diamidino-2-phenylindole. scRNA-seq analysis was performed as described previously.10,21 Single cells were individually sorted by fluorescence-activated cell sorting into wells of a 96-well polymerase chain reaction plate containing lysis buffer. The Illumina Nextera XT DNA preparation kit was used to prepare libraries. Pooled libraries were sequenced by using the Illumina HiSequation 2500 system and re-sequenced by using the Illumina HiSequation 4000 system (single-end 125 bp reads). Reads were aligned using G-SNAP,22 and the mapped reads were assigned to Ensembl genes (release 81)23 by HTSeq.24
To pass quality control, cells were required to have at least 200 000 reads mapping to nuclear genes, at least 4000 genes detected, less than 10% of mapped reads mapping to mitochondrial genes, and less than 50% of mapped reads mapping to the External RNA Controls Consortium (ERCC) spike-ins (#4456740, Life Technologies) (supplemental Figure 1, available on the Blood Web site). Reads were normalized by following the method of Lun et al25 using an initial clustering step to group cells with similar expression patterns. ERCC spike-ins were used to estimate the level of technical variance as described by Brennecke et al.26 Variable genes were defined as having a squared coefficient of variation exceeding technical noise, with 4773 genes passing this threshold (supplemental Figure 2B).
Raw data has been uploaded to National Center for Biotechnology Information GEO (accession number GSE81682). Index data were normalized in R (https://www.r-project.org), using flowCore to extract and compensate the data and ComBat from the sva package to normalize the data. Thresholds for each population were assigned retrospectively based on published literature27-30 and compared with normalized index data with FlowJo (Treestar). E-SLAM (CD48–CD150+CD45+EPCR+) cells were gated as EPCR+CD48–CD150+ because CD45 was not available in the index data. The gates were set to either cover all cells (broad gating) or leave unclassified cells in between populations to ensure that the gates did not contain any overlap (narrow gating).
Computational analysis
All computational analysis was performed in the R programming environment (https://www.r-project.org). Hierarchical clustering was performed by using the hclust function, with distance (1 – Spearman’s correlation)/2 and average linkage. Discrete clusters were identified by using cutreeDynamic (dynamicTreeCut package), with the hybrid method and minimum cluster size of 10. The deepSplit parameter was set to 1, resulting in 4 broad clusters. For each cluster, gene expression was compared between cells in the cluster and the rest of the data set. Genes expressed (log2 expression value >4) in at least half the cells in a cluster were tested for differential expression by using a Wilcoxon rank sum test with Benjamini-Hochberg correction. Genes with a false discovery rate <0.001 were ranked by fold change, and the 10 genes with highest fold change for each cluster are displayed in Figure 1B.
Generating linked transcriptional and surface marker profiles for more than 1600 single HSPCs. (A) Schematic of the sorting strategy that was used paired with index sorting data. Bone marrow cells were stained with 9 antibodies against various cell surface markers to isolate HSPCs (Lin−c-Kit+Sca1+ [L−S+K+]) and progenitors (Lin−c-Kit+Sca1− [L−S−K+]). Almost all cells in the Flk2-CD34 gate and the CD16/32-Flk2 gate were collected for HSPCs and progenitors, respectively, within broad, all-encompassing gates. In addition, LT-HSCs (Lin–c-Kit+Sca1+CD34–Flk2–) were collected separately to ensure that adequate numbers were collected. Each cell population retrospectively identified is shown in the table; colors and names remain consistent throughout the text. Letters indicate populations in the flow cytometry diagrams. (B) Unsupervised hierarchical clustering of gene expression data for all cells. Clustering was performed by using all 4773 variable genes except Ly6a/Sca-1 to avoid bias in clustering. The cells split into 4 major clusters (cluster 1, purple; cluster 2, turquoise; cluster 3, gold; cluster 4, pink). The top 10 genes enriched in each cluster are displayed in the heat map, showing gene expression on a log2 scale from blue to orange (low to high). The clusters were also compared by cell type composition, following both broad and narrow gating strategies. Broad gating involved the classification of all cells into a cell type category, whereas narrow gating included only cells that are more likely to fit the predefined HSPC classification, gated around the greatest density of cells within the population gating strategy. Cell type is colored on the basis of the scheme used in Figure 1A. Gray cells in the narrow gating strategy represent cells unassigned to any population. FACS, fluorescence-activated cell sorting; FSC-H, forward-scattered light-height; Prog, progenitor.
Generating linked transcriptional and surface marker profiles for more than 1600 single HSPCs. (A) Schematic of the sorting strategy that was used paired with index sorting data. Bone marrow cells were stained with 9 antibodies against various cell surface markers to isolate HSPCs (Lin−c-Kit+Sca1+ [L−S+K+]) and progenitors (Lin−c-Kit+Sca1− [L−S−K+]). Almost all cells in the Flk2-CD34 gate and the CD16/32-Flk2 gate were collected for HSPCs and progenitors, respectively, within broad, all-encompassing gates. In addition, LT-HSCs (Lin–c-Kit+Sca1+CD34–Flk2–) were collected separately to ensure that adequate numbers were collected. Each cell population retrospectively identified is shown in the table; colors and names remain consistent throughout the text. Letters indicate populations in the flow cytometry diagrams. (B) Unsupervised hierarchical clustering of gene expression data for all cells. Clustering was performed by using all 4773 variable genes except Ly6a/Sca-1 to avoid bias in clustering. The cells split into 4 major clusters (cluster 1, purple; cluster 2, turquoise; cluster 3, gold; cluster 4, pink). The top 10 genes enriched in each cluster are displayed in the heat map, showing gene expression on a log2 scale from blue to orange (low to high). The clusters were also compared by cell type composition, following both broad and narrow gating strategies. Broad gating involved the classification of all cells into a cell type category, whereas narrow gating included only cells that are more likely to fit the predefined HSPC classification, gated around the greatest density of cells within the population gating strategy. Cell type is colored on the basis of the scheme used in Figure 1A. Gray cells in the narrow gating strategy represent cells unassigned to any population. FACS, fluorescence-activated cell sorting; FSC-H, forward-scattered light-height; Prog, progenitor.
Dimensionality reduction was performed on log2-transformed expression data for the 4773 variable genes by using the diffusion map method31 (destiny package32 ) with cosine distance and Gaussian kernel width of 0.16. Three-dimensional plots were produced by using the scatter3D function from the plot3D package, and the dm.predict function was used to project external data. Because of high cell numbers, data from Kowalczyk et al33 were randomly sampled to obtain 50 cells from each condition (cell type, condition, and strain) for clearer visualization.
Three-dimensional diffusion map embedding was used to identify a start cell (within the E-SLAM population) and end cells for each of the 3 lineages (E, erythroid; GM, granulocyte-macrophage; and L, lymphoid). Identifying broad branches between start and end cells was done by finding cells centered around the shortest paths in the diffusion map, following the procedure of Ocone et al.13 To identify genes upregulated or downregulated with trajectories, cells were ordered in pseudotime, and gene expression was smoothed by calculating the mean for a sliding window of size 20. Spearman’s correlation between smoothed pseudotime and expression values was calculated for each gene, and genes with absolute correlation >0.5 were identified and clustered by using hierarchical clustering with average linkage on Spearman’s correlation.
Gene set enrichment analysis was performed in Enrichr.34 Results with adjusted P value <.05 (using Benjamini-Hochberg correction for multiple testing) were considered significant. Full tables of results can be found in the supplemental Data. Cell cycle genes were downloaded from Reactome (http://www.reactome.org/). Cell cycle category was inferred by using a recently described method.35 To estimate absolute gene expression, external ERCC spike-ins were used to normalize reads within each plate by calculating spike-in size factors using the computeSpikeFactors function from the scran package before normalizing cells with these size factors. To account for batch effect differences in ERCC concentration between lanes (supplemental Figure 5), we applied ComBat from the SVA package, using the sorting gate (HSPC/Prog/LT-HSC) as an adjustment variable. Estimates of the total RNA content were calculated by summing absolute normalized counts per cell. Significance of differences in RNA content and forward-scattered light-height between cell types was calculated by using a 1-way analysis of variance test. To identify genes downregulated in pseudotime in absolute terms, the previously obtained downregulated lists (found by using relative gene expression values) were filtered to remove any genes that did not have a greater than twofold absolute expression change between the first 10% of cells in a pseudotime trajectory and the final 10%.
Results
An atlas of single-cell HSPC expression profiles
Single-cell resolution RNA-Seq of embryonic stem and muscle progenitor cell differentiation has demonstrated that differentiation likely occurs as a near-continuous process, with gradual changes in gene expression as cells traverse the transcriptional landscape.14,36 To comprehensively sample cells across the entire spectrum of the mouse HSPC transcriptional landscape, we isolated single cells by using two broad sorting gates based on c-Kit and Sca1 protein expression, encompassing long-term HSCs (LT-HSCs; Lin–c-Kit+Sca1+CD34–Flk2–), lymphoid multipotent progenitors (LMPPs), and multipotent progenitors (MPPs) in one gate called the HSPC gate, and megakaryocyte-erythrocyte progenitors (MEPs), common myeloid progenitors (CMPs), and granulocyte-monocyte progenitors (GMPs) in the second gate called the Progenitor/Prog gate (Figure 1A). Because LT-HSCs are much less frequent than other populations in the HSPC gate, additional LT-HSCs were also sorted. Cells were retrospectively categorized into specific HSPC populations27,28 by using index-sorting data.10 Each cell was also stained with 3 additional antibodies against CD150, CD48, and EPCR to retrospectively assign cells to other commonly used sorting schemes for populations such as E-SLAM3 or MPP subpopulations.27,29
Single cells were processed for RNA-Seq as described21 with 156 HSCs, 701 HSPCs, and 799 progenitors passing stringent quality control parameters (see “Methods”). Technical noise analysis26 revealed 4773 genes with expression variability exceeding technical noise. Unsupervised clustering partitioned the 1656 cells into 4 major clusters (Figure 1B). Cluster 1 is mostly made up of LT-HSCs and is represented by genes such as Procr (EPCR) and Trpc6. Clusters 2 and 3 are both composed of all investigated cell types and share expression of many of the representative genes but are differentiated by higher expression of several genes, including Ccl9, Clec12a, and Tyrobp in cluster 3. Cluster 4 is mainly composed of MEPs and is characterized by expression of genes such as hemoglobin alpha, adult chain 1 (Hba-a1) and Smim1. This analysis suggests that the transcriptomes of 1656 single HSPCs presented here provide new opportunities for exploring the transcriptional landscape of early HSC differentiation at single-cell resolution.
Visualizing gene expression along the continuum of HSPC differentiation
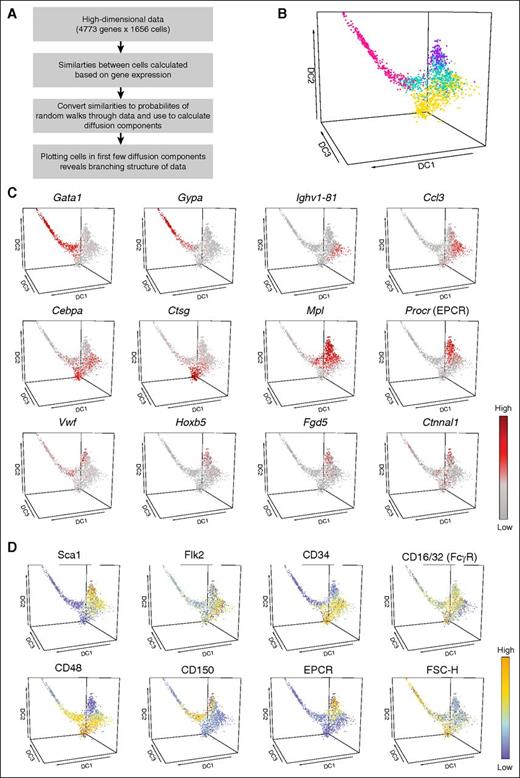
Diffusion maps have recently emerged as a dimensionality reduction procedure particularly suited to displaying continuous differentiation processes from single-cell snapshot data.11,31,37 When applied to the 1656 cells profiled here (Figure 2A), an intuitive graphical representation of the early process of HSPC differentiation emerges. The diffusion map can be colored on the basis of the previously identified clusters (Figure 2B), revealing that clusters 1 (purple), 3 (gold), and 4 (pink) form separate branches of the diffusion map, and cluster 2 (turquoise) encompasses cells among the 3 branches. Expression levels of individual genes can be plotted in the diffusion map to reveal their expression profiles across the HSPC transcriptional landscape (Figure 2C). Gata1 expression is concentrated in cluster 4, consistent with it being made up mostly of MEPs. Procr and Mpl expression is seen mainly in cluster 1, which is made up of LT-HSCs. Of note, the recently reported LT-HSC markers Hoxb5, Fgd5, and Ctnaal1/α-catulin38-40 all showed predominant expression in cluster 1.
Multidimensional analysis can be used to visualize gene expression across HSPC differentiation. (A) Schematic explaining how diffusion maps are used as a dimensionality reduction procedure. (B) Diffusion map of all cells was colored on the basis of previously defined clusters (cluster 1, purple; cluster 2, turquoise; cluster 3, gold; cluster 4, pink). Diffusion components 1, 2, and 3 are shown. (C) Diffusion map of all cells was colored according to the expression of selected genes. The genes were chosen on the basis of published literature or were identified computationally as highly expressed in specific cell populations. The color corresponds to a log2 scale of expression ranging between 0 and the maximum value for each gene. (D) Diffusion map of all cells was colored by surface marker expression from the normalized index data. The majority of these markers were used for cell selection, with the exception of CD48, CD150, and EPCR. The color corresponds to a linear scale of expression ranging between the minimum and maximum value for each marker.
Multidimensional analysis can be used to visualize gene expression across HSPC differentiation. (A) Schematic explaining how diffusion maps are used as a dimensionality reduction procedure. (B) Diffusion map of all cells was colored on the basis of previously defined clusters (cluster 1, purple; cluster 2, turquoise; cluster 3, gold; cluster 4, pink). Diffusion components 1, 2, and 3 are shown. (C) Diffusion map of all cells was colored according to the expression of selected genes. The genes were chosen on the basis of published literature or were identified computationally as highly expressed in specific cell populations. The color corresponds to a log2 scale of expression ranging between 0 and the maximum value for each gene. (D) Diffusion map of all cells was colored by surface marker expression from the normalized index data. The majority of these markers were used for cell selection, with the exception of CD48, CD150, and EPCR. The color corresponds to a linear scale of expression ranging between the minimum and maximum value for each marker.
Visualization of surface marker expression from the normalized index data marked coherent territories within the diffusion map consistent with a robust separation of HSCs and more mature progenitors (Figure 2D). These results illustrate how the diffusion map representation of our data set is a powerful way of interrogating the gene expression of any gene across the transcriptional landscape of HSPC differentiation. We therefore developed a user-friendly Web site (http://blood.stemcells.cam.ac.uk/single_cell_atlas.html) where users can explore the three-dimensional structure of the diffusion map graph as well as visualize expression profiles for any gene of interest and surface marker expression. Of note, alternative dimensionality reduction methods such as principal component analysis showed similar relationships between the clusters (supplemental Figure 4). This novel data set and accompanying online resource permits interrogation of individual genes and surface markers at single-cell resolution and can be broadly applied to a range of applications, including full integration of other single-cell data sets.
The single-cell transcriptional landscape illustrates the nature of HSPC populations and cellular phenotypes
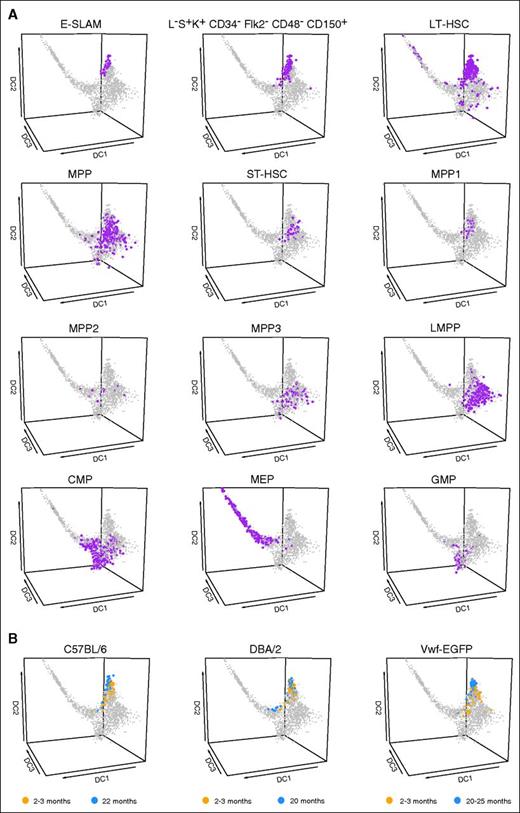
The relationships between different surface-marker-defined HSPC populations remain an area of active debate. After a uniform panel of 9 surface markers was used for index sorting, cells were retrospectively assigned to 12 distinct HSPC phenotypes and displayed in the diffusion map (Figure 3A). With the exception of the CMP population, which has been described as functionally heterogeneous,41 all other populations occupied defined territories. The original article describing MEPs showed that GMPs are more common than MEPs42 ; however, they performed partial lineage depletion, which differs from the conditions used in this study, thus influencing the ratios of GMPs, MEPs, and CMPs isolated. Importantly, although lineage depletion can be variable, retrospective back-gating places the cells accurately. The 3 populations containing LT-HSCs overlapped as expected, with additional substantial overlaps between MPP3 and LMPP, and potential progressions such as a putative journey from E-SLAM via short-term HSCs (ST-HSCs) and LMPP to GMP.
The single-cell HSPC transcriptional landscape can be used to visualize HSPC populations and their relationships. (A) Diffusion map of all cells was colored on the basis of cell population using narrow gating. All populations were identified retrospectively by using the index sorting data. Populations were identified by using normalized index data. The cells of interest for each population are colored purple and enlarged for easier visibility. (B) Diffusion map of all cells with projection of data from recently published data sets. Data collected by Kowalczyk et al (C57BL/6, DBA/2) and Grover et al (Vwf-EGFP) is displayed. Both groups collected HSCs from mice 2 to 3 months (orange) and 20 to 25 months (blue) old. HSCs were defined as Lin–c-Kit+Sca1+CD150+CD48–.
The single-cell HSPC transcriptional landscape can be used to visualize HSPC populations and their relationships. (A) Diffusion map of all cells was colored on the basis of cell population using narrow gating. All populations were identified retrospectively by using the index sorting data. Populations were identified by using normalized index data. The cells of interest for each population are colored purple and enlarged for easier visibility. (B) Diffusion map of all cells with projection of data from recently published data sets. Data collected by Kowalczyk et al (C57BL/6, DBA/2) and Grover et al (Vwf-EGFP) is displayed. Both groups collected HSCs from mice 2 to 3 months (orange) and 20 to 25 months (blue) old. HSCs were defined as Lin–c-Kit+Sca1+CD150+CD48–.
The diffusion map protocol has recently been developed to permit projection of new data into the coordinates of an existing diffusion map,32 which allowed us to interrogate cellular phenotypes of other recently published single-cell data sets. Projection of young and old HSCs in C57BL/6 and DBA/2 mouse strains43 and Vwf-EGFP mice33 showed that both young and old HSCs cluster together with LT-HSCs from our data set, with old HSCs forming a tighter cluster suggestive of a more homogeneous population. Therefore, this analysis not only demonstrates that our large expression atlas permits robust comparisons between single-cell data sets generated in different labs, it also reveals a consistent phenotypic change of old HSCs in both studies, in which old stem cells are more concentrated in what seems to be the core HSC territory of the diffusion map.
Mapping differentiation trajectories from the single-cell expression landscape
Having established that single cells in the diffusion map are arranged in a pattern consistent with known lineage relationships, we next identified 3 differentiation trajectories (see “Methods”) starting each time with E-SLAM HSCs and ending with E, GM, and L progenitors (Figure 4A). On the basis of gene expression profiles, each cell within a differentiation trajectory is given a pseudotime timestamp and can therefore be arranged in a pseudotemporal ordering (see “Methods”). Visualization of surface marker expression from the index data revealed dynamic profiles consistent with known expression patterns, thus validating the pseudotemporal ordering (Figure 4B). This analysis also showed that the E trajectory traverses through a significant proportion of cells co-expressing CD150 and CD48, whereas the proportion of cells with that surface marker phenotype is much smaller for the GM and L trajectories.
Pseudotime analysis reveals trends in surface marker and gene expression for differentiation trajectories. (A) Diffusion map colored by pseudotime trajectories to E, GM, and L fates. Each trajectory starts from an HSC (blue) and ends with a progenitor (red). (B) Changes in surface marker expression and FSC-H through pseudotime for each of the 3 trajectories obtained from the normalized index data. For each trajectory, it is possible to see what cell types are passed through to reach the final cell fate. (C) Normalized expression of genes positively (up) or negatively (down) correlated with the pseudotemporal ordering for each trajectory. Mean normalized expression is plotted with standard deviation. (D) Most significant relevant terms from gene set enrichment analysis for all the trajectories, performed in Enrichr. Terms with an adjusted P value <.05 (using Benjamini-Hochberg correction for multiple testing) were considered significant. The full tables of results can be found in the supplemental Data. MGI, mouse genome informatics.
Pseudotime analysis reveals trends in surface marker and gene expression for differentiation trajectories. (A) Diffusion map colored by pseudotime trajectories to E, GM, and L fates. Each trajectory starts from an HSC (blue) and ends with a progenitor (red). (B) Changes in surface marker expression and FSC-H through pseudotime for each of the 3 trajectories obtained from the normalized index data. For each trajectory, it is possible to see what cell types are passed through to reach the final cell fate. (C) Normalized expression of genes positively (up) or negatively (down) correlated with the pseudotemporal ordering for each trajectory. Mean normalized expression is plotted with standard deviation. (D) Most significant relevant terms from gene set enrichment analysis for all the trajectories, performed in Enrichr. Terms with an adjusted P value <.05 (using Benjamini-Hochberg correction for multiple testing) were considered significant. The full tables of results can be found in the supplemental Data. MGI, mouse genome informatics.
We next identified genes showing statistically significant positive or negative correlation with the pseudotemporal ordering (Figure 4C). Gene set enrichment analysis (Figure 4D) showed enrichments consistent with the respective trajectories such as tetrapyrrole biosynthesis for E upregulated genes and neutrophil-mediated immunity for GM upregulated genes. This analysis also revealed a major contribution of cell cycle–associated genes to both the E and GM upregulated genes. The 3 differentiation trajectories mapped out here are therefore consistent with current knowledge of early hematopoiesis, suggesting that the pseudotime reconstruction will provide a powerful means of charting the dynamic processes that underlie early HSPC differentiation at single-cell resolution.
Single-cell resolution analysis of cell cycle activation during HSPC differentiation
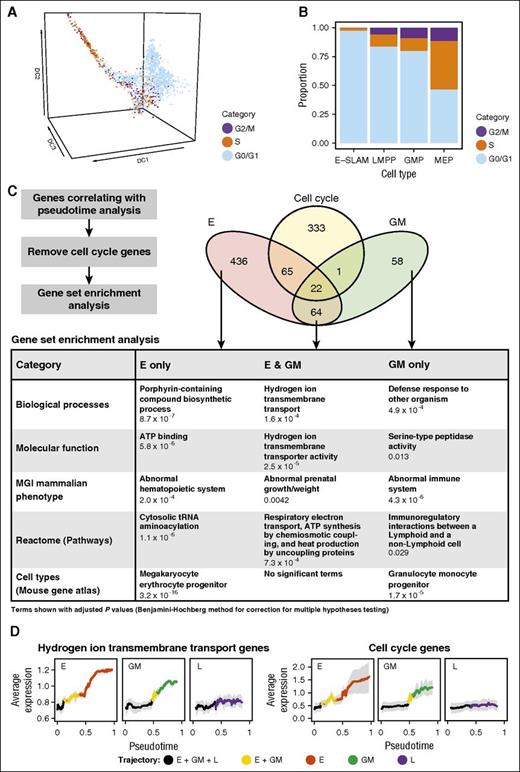
Having identified cell cycle as the most highly enriched term for the genes upregulated along both the E and GM trajectories, we next took advantage of a recently reported predictor for allocating individual cells to G0/G1, S, and G2/M cell cycle categories based on their single-cell transcriptomes.35 The distribution of single cells across these 3 cell cycle categories was in good agreement with the enrichment of cell cycle terms in the genes upregulated along the E and GM trajectories (Figure 5A-B). The analysis also demonstrated that large-scale transitioning of cells to S and G2/M phase occurs after the divergence of the L trajectory from the E and GM trajectories, thus suggesting that transition to rapid cell cycling is secondary to transcriptional diversification.
Analysis of cell cycle activation during HSPC differentiation at single-cell resolution. (A) Diffusion map of all cells colored by computationally assigned cell cycle category. There is no assignment for G0 separately because of limitations of the method. (B) Proportion of E-SLAMs, LMPPs, GMPs, and MEPs in each of the cell cycle categories. The cell types displayed are based on the narrow gating strategy. (C) Gene set enrichment analysis was performed for the 3 trajectories after the removal of cell cycle genes. The most relevant significant terms for genes positively correlated with pseudotime analysis are shown. Terms with an adjusted P value <.05 (using Benjamini-Hochberg correction for multiple testing) were considered significant. The full tables of results can be found in the supplemental Data. (D) Average expression of hydrogen ion transmembrane transport genes and cell cycle genes across pseudotime. Each gene was normalized across the median of all 3 trajectories for plotting. The average expression is colored by trajectory, and means are shown with standard deviations. tRNA, transfer RNA.
Analysis of cell cycle activation during HSPC differentiation at single-cell resolution. (A) Diffusion map of all cells colored by computationally assigned cell cycle category. There is no assignment for G0 separately because of limitations of the method. (B) Proportion of E-SLAMs, LMPPs, GMPs, and MEPs in each of the cell cycle categories. The cell types displayed are based on the narrow gating strategy. (C) Gene set enrichment analysis was performed for the 3 trajectories after the removal of cell cycle genes. The most relevant significant terms for genes positively correlated with pseudotime analysis are shown. Terms with an adjusted P value <.05 (using Benjamini-Hochberg correction for multiple testing) were considered significant. The full tables of results can be found in the supplemental Data. (D) Average expression of hydrogen ion transmembrane transport genes and cell cycle genes across pseudotime. Each gene was normalized across the median of all 3 trajectories for plotting. The average expression is colored by trajectory, and means are shown with standard deviations. tRNA, transfer RNA.
Because terms associated with cell cycle had dominated the gene set enrichment analysis for the E and GM trajectories described in Figure 4, we next intersected the E and GM upregulated genes with a curated set of 405 cell cycle–associated genes. The filtered E-only and GM-only gene sets showed strong enrichment for terms associated with their known biological functions, such as porphyrin biosynthesis for heme production (E only) and defense response to other organisms (GM only) (Figure 5C). Of note, the cell cycle–filtered genes upregulated in both the E and GM trajectories showed strong enrichment for terms associated with mitochondrial adenosine triphosphate production, consistent with previous reports that HSCs primarily use glycolysis44-46 but switch to mitochondrial oxidative phosphorylation to meet the rapidly increasing energy demands for differentiation.47
We next investigated how hydrogen ion transmembrane transport gene and cell cycle gene expression changes through pseudotime (Figure 5D). In the GM trajectory, expression increases after cells enter the GM/E trajectory, with highest expression achieved once the cells enter the GM-only trajectory. For the E trajectory, expression already increases before cells leave the GM/E/L trajectory and continues to increase as cells transition into the E trajectory. As expected from the gene set enrichment analysis (Figure 4D), there is no substantial increase of either hydrogen ion transmembrane transport or cell cycle genes along the L trajectory.
Identification of genes downregulated in absolute terms during HSC differentiation
The relative quiescence and low metabolic activity of HSCs might be reflected in low amounts of total messenger RNA (mRNA) per cell. However, conventional bulk microarray or RNA-Seq analysis is geared toward identifying relative expression differences only. Conversely, single-cell profiling can be used to estimate absolute differences in total mRNA content. To estimate total mRNA content per cell, we used external spike-in controls, sorted single cells from HSPC, progenitor, and LT-HSC gates into all twenty 96-well plates in a predetermined layout, and sequenced each plate on a single lane so that consistent differences between the amounts of reads between cell types would become detectable (Figure 6A). Estimation of absolute mRNA content per cell revealed a gradual increase in average mRNA content from E-SLAM HSCs to LMPPs to GMPs to MEPs (Figure 6B-C) (cells assigned to populations based on index sorting data; Figure 3). Of note, forward scatter is recognized as a correlate to cell size and showed a similar, but not identical, pattern (Figure 6D), thus suggesting that mRNA content per cell is related, but not completely coupled, to cell size during early HSC differentiation.
Single-cell analysis can be used to estimate absolute differences in total mRNA content across cell types. (A) Schematic explanation of how plate composition and ERCC spike-ins are used to estimate absolute RNA levels. The plate organization for this study included cells from multiple sorting gates (HSPC, Prog, LT-HSCs) and each well contained ERCC spike-ins. The sequencing depth varies across lanes and cell types; therefore, ERCC spike-ins are used to normalize across cell types within a lane, in which the spike-in content becomes level within a lane but cell mRNA content may still vary. After this step, RNA content can be normalized across lanes. (B) Diffusion map of all cells was colored by RNA content. Estimates of total RNA content were calculated by summing the absolute normalized counts per cell. The scale ranges from blue to green to yellow to red with increasing RNA content. (C) Sum of normalized counts for E-SLAMs, LMPPs, GMPs, and MEPs colored by the scheme used in Figure 1A. Significance in differences in RNA content between cell types was calculated by using a 1-way analysis of variance test (**P < .001; ***P < .0001). (D) FSC-H for E-SLAMs, LMPPs, GMPs, and MEPs, colored by the scheme used in Figure 1A. FSC-H is used as an indicator of cell size. Significance in differences in FSC-H between cell types was calculated by using a 1-way analysis of variance test (**P < .001; ***P < .0001). (E) Most relevant significant terms from gene enrichment expression analysis on genes downregulated in absolute terms in E-only, GM-only, and E and GM trajectories. The numbers of genes showing downregulation along pseudotime in absolute terms is displayed in the Venn diagram. Terms with an adjusted P value <.05 (using Benjamini-Hochberg correction for multiple testing) were considered significant. The full tables of results can be found in the supplemental Data.
Single-cell analysis can be used to estimate absolute differences in total mRNA content across cell types. (A) Schematic explanation of how plate composition and ERCC spike-ins are used to estimate absolute RNA levels. The plate organization for this study included cells from multiple sorting gates (HSPC, Prog, LT-HSCs) and each well contained ERCC spike-ins. The sequencing depth varies across lanes and cell types; therefore, ERCC spike-ins are used to normalize across cell types within a lane, in which the spike-in content becomes level within a lane but cell mRNA content may still vary. After this step, RNA content can be normalized across lanes. (B) Diffusion map of all cells was colored by RNA content. Estimates of total RNA content were calculated by summing the absolute normalized counts per cell. The scale ranges from blue to green to yellow to red with increasing RNA content. (C) Sum of normalized counts for E-SLAMs, LMPPs, GMPs, and MEPs colored by the scheme used in Figure 1A. Significance in differences in RNA content between cell types was calculated by using a 1-way analysis of variance test (**P < .001; ***P < .0001). (D) FSC-H for E-SLAMs, LMPPs, GMPs, and MEPs, colored by the scheme used in Figure 1A. FSC-H is used as an indicator of cell size. Significance in differences in FSC-H between cell types was calculated by using a 1-way analysis of variance test (**P < .001; ***P < .0001). (E) Most relevant significant terms from gene enrichment expression analysis on genes downregulated in absolute terms in E-only, GM-only, and E and GM trajectories. The numbers of genes showing downregulation along pseudotime in absolute terms is displayed in the Venn diagram. Terms with an adjusted P value <.05 (using Benjamini-Hochberg correction for multiple testing) were considered significant. The full tables of results can be found in the supplemental Data.
We next used the spike-in based normalization to investigate whether genes identified as downregulated in Figure 4 were indeed downregulated in real terms (eg, fewer mRNA molecules per single cell). Importantly, conventional analysis would not have been able to distinguish this absolute downregulation from relative downregulation. In a situation in which there is an increase of total amount of RNA per cell, as our spike-in based analysis shows for HSC differentiation, a given gene might appear to be downregulated in the relative expression analysis whereas it actually stays the same in absolute terms while a large fraction of the transcriptome is upregulated. However, the majority of downregulated genes from Figure 4 were downregulated in absolute terms along the E and GM trajectories (109 of 112 for E and 55 of 56 for GM), thus highlighting a subset of genes actively expressed in HSCs despite their quiescent and metabolically less-active state (supplemental Table 1). Gene set enrichment analysis showed enrichment for terms associated with megakaryocytes, although on closer inspection, this corresponded to genes such as Mpl and Procr, known to be highly expressed in HSCs. Only 18 genes were specifically downregulated in the GM trajectory, thus precluding the identification of any statistically significant gene set overlaps. Terms enriched with the E downregulated genes corresponded to genes associated with the immune response. Taken together, these data demonstrate that single-cell analysis allows estimation of total mRNA amounts per cell in the various HSPC compartments, thus allowing identification of genes that are, in real terms, more highly expressed in HSCs than the various downstream progenitors such as GMP and MEP.
Discussion
Here we have taken advantage of recent advances in molecular profiling technologies to provide a single-cell resolution expression atlas of early blood stem cell differentiation, which (1) overcomes several shortcomings of population-based bulk expression profiling, (2) provides new insights into the diversification of transcriptional programs during HSC differentiation, and (3) represents a powerful new resource for the hematopoiesis research community facilitated by the development of a new user-friendly Web site.
Previous bulk transcriptome analyses have made several important contributions to enhancing our understanding of HSPCs, including the identification of new candidate regulators48 and complex patterns of coordinately expressed gene sets.16 Comprehensive single-cell transcriptome data provide opportunities not readily available with conventional population-average data. For example, absolute differences in mRNA levels can be estimated for cells belonging to distinct differentiation stages. The quiescent nature27 and low metabolic activity46,47 of HSCs might have been taken to imply that the HSC state is characterized by a general low level of transcription, in line with the well-documented low activity of Myc in HSCs.49-51 Our data confirm this hypothesis in some respects by demonstrating that HSCs consistently contain less mRNA per cell than E and GM cells. Nevertheless, there exists a subset of genes with higher expression in absolute terms in HSCs, suggesting that some genes might contribute to actively maintaining the stem cell state.
The ability to project external single-cell transcriptional data onto our single-cell transcriptome atlas offers an attractive method of hypothesis generation. We projected data from 2 different laboratories and 2 different mouse strains33,43 that all gave similar results, thus underscoring the robustness of this approach. When compared with HSCs from young mice, HSCs from old mice were more confined to the HSC territory of the diffusion map, suggesting that HSCs from old mice represent a more molecularly homogeneous population, with fewer cells already engaged in a differentiation trajectory. Of note, this observation was not reported in the 2 original publications, presumably because they lacked the extensive landscape of single HSPC transcriptional states as a comparator. Interestingly, however, conventional expression profiling of HSCs from old mice when coupled with epigenetic analysis had already suggested that in old HSCs, the transcriptomic and epigenetic landscape promotes HSC self-renewal at the expense of differentiation.52 Future exploitations of the single-cell atlas as a comparator are likely to include the analysis of single-cell transcriptomes from mouse models, including inducible mouse models of leukemia.
When gene expression states are measured by using thousands of genes, progression of a cell through a differentiation program can be thought of as a journey through a transcriptional landscape. This study captures 1656 single-cell gene expression snapshots of the HSPC transcriptional landscape, which provide several important insights. For example, dimensionality reduction methods such as diffusion maps represent a useful way to visualize and interpret data sets of more than 8 million data points (eg, 1656 cells × 4773 heterogeneously expressed genes). This is supported by the observation that previously defined HSPC populations form coherent groupings on the diffusion map with one major exception—CMPs, which have recently been described as highly heterogeneous.41,53
Furthermore, although the arrangement of cells in the diffusion map is consistent with known developmental progressions (eg, LT-HSC to ST-HSC to LMPP to GMP), there is substantial intermingling within transition zones. Some cells sorted as LMPPs, for example, will therefore be virtually identical at the transcriptome level to cells sorted as ST-HSCs. Moreover, for other transitions such as LMPP to GMP, conventional gating fails to capture a substantial number of cells in the transition zone. Of note, molecular characterization of such transition cells may be particularly important to advance our understanding of cellular differentiation.
A number of methods have been developed to reconstruct differentiation trajectories from single-cell expression data.8,14 Given the likely plasticity of immature cells, we opted for developing broad trajectories in which a given cell at any moment in time would have the option of making sideway movements rather than just finding the shortest path between the 2 end points. It is remarkable, therefore, that even with these relatively broad trajectories, the 3 journeys reconstructed here already diverge within the part of the diffusion map occupied mostly by the ST-HSC population. Although this observation is at odds with the more traditional view of the hematopoietic lineage tree,54 it is consistent with recent analysis of both mouse and human cell fate diversification.41,53,55 Importantly, we now provide for the first time a reconstruction of the likely dynamics of expression changes during these early stages of HSPC fate diversification.
An important consideration with single-cell RNA-Seq is to strike a balance between the number of cells profiled and the sequencing depth achieved for each cell. We opted for substantial sequencing depth, detecting on average 6558 protein-coding genes per cell. Emerging droplet sequencing technology facilitates increased throughput,36 but current methods do not afford ways of recording surface marker expression analogous to the index sorting used here. Moreover, studies published so far have opted for much lower sequencing depth to keep overall costs manageable. However, this makes it impossible to develop an online resource such as the one reported here, which can be used to display the expression profile for any gene of interest. Substantial sequencing depth is also required if single-cell data are to be exploited for the discovery of molecular mechanisms that may drive cellular differentiation and diversification. The data set and analysis reported here should be well placed to serve this function for the wider hematopoiesis research community.
This article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Reiner Schulte, Chiara Cossetti, and Michal Maj at the Cambridge Institute for Medical Research Flow Cytometry Core for their help with cell sorting, Dean Pask and Tina Hamilton for technical assistance, and Wajid Jawaid, Paul Sumption, and Chee Lim for help with setting up the Web site.
This work was supported by grants from Bloodwise, Cancer Research UK, Biotechnology and Biological Sciences Research Council, Leukemia Lymphoma Society, the National Institute for Health Research Cambridge Biomedical Research Centre, and core support grants by Wellcome Trust to the Cambridge Institute for Medical Research and Wellcome Trust-Medical Research Council Cambridge Stem Cell Institute. S.N. and F.K.H. are recipients of Medical Research Council PhD studentships. D.G.K. is the recipient of a Bennett Fellowship from Bloodwise, and E.L. is the recipient of a Sir Henry Dale Fellowship from the Wellcome Trust.
Authorship
Contribution: S.N., M.S., N.K.W., and D.G.K. performed experiments; F.K.H. analyzed single-cell sequencing data; F.K.H. and B.P.S. analyzed index data; E.D. mapped sequencing data; E.L., N.K.W., D.G.K., and B.G. designed and supervised the study; and S.N., F.K.H., E.L., N.K.W., D.G.K., and B.G. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Berthold Göttgens, University of Cambridge, Cambridge Institute for Medical Research, Hills Rd, Cambridge CB2 0XY, United Kingdom; e-mail: bg200@cam.ac.uk; David G. Kent, University of Cambridge, Cambridge Institute for Medical Research, Hills Rd, Cambridge CB2 0XY, United Kingdom; e-mail: dgk23@cam.ac.uk; and Nicola K. Wilson, University of Cambridge, Cambridge Institute for Medical Research, Hills Rd, Cambridge CB2 0XY, United Kingdom; e-mail: nkw22@cam.ac.uk.
References
Author notes
S.N. and F.K.H. contributed equally to this study.

![Figure 1. Generating linked transcriptional and surface marker profiles for more than 1600 single HSPCs. (A) Schematic of the sorting strategy that was used paired with index sorting data. Bone marrow cells were stained with 9 antibodies against various cell surface markers to isolate HSPCs (Lin−c-Kit+Sca1+ [L−S+K+]) and progenitors (Lin−c-Kit+Sca1− [L−S−K+]). Almost all cells in the Flk2-CD34 gate and the CD16/32-Flk2 gate were collected for HSPCs and progenitors, respectively, within broad, all-encompassing gates. In addition, LT-HSCs (Lin–c-Kit+Sca1+CD34–Flk2–) were collected separately to ensure that adequate numbers were collected. Each cell population retrospectively identified is shown in the table; colors and names remain consistent throughout the text. Letters indicate populations in the flow cytometry diagrams. (B) Unsupervised hierarchical clustering of gene expression data for all cells. Clustering was performed by using all 4773 variable genes except Ly6a/Sca-1 to avoid bias in clustering. The cells split into 4 major clusters (cluster 1, purple; cluster 2, turquoise; cluster 3, gold; cluster 4, pink). The top 10 genes enriched in each cluster are displayed in the heat map, showing gene expression on a log2 scale from blue to orange (low to high). The clusters were also compared by cell type composition, following both broad and narrow gating strategies. Broad gating involved the classification of all cells into a cell type category, whereas narrow gating included only cells that are more likely to fit the predefined HSPC classification, gated around the greatest density of cells within the population gating strategy. Cell type is colored on the basis of the scheme used in Figure 1A. Gray cells in the narrow gating strategy represent cells unassigned to any population. FACS, fluorescence-activated cell sorting; FSC-H, forward-scattered light-height; Prog, progenitor.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/128/8/10.1182_blood-2016-05-716480/4/m_e20f1.jpeg?Expires=1763466278&Signature=4zpSAeo8DDnkUNUsuAVnlvJ49daXJNBw~5wqOBjZTVNY7OVGzlDffk~FG76ZeNtilhi8k6~BAT1KoR1vcRmBhq2Hg2xkm7itH7dh351U7F-AGADkNffTa1mNqziIoNA10ZNahVIyclG0Y8jG0BLwdguvMc-ZhjzTJ5GtafW1~D-oaC1Qy31CqTIAduKaX6hQl6-CLRdJ7HeoO85d9owJmEYAW5CbLLcDd9ZO6uQAKXszO78pvEyf-cpDk3rmR-YeaTVBv2IFMwEt3OL4CFI94QGeNQQHlthHNdHS3JPAfOaeM4E2HFyaHX8QRkXX0cpBHqZHUH0a15K4ayC5b0n9Pg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)