Key Points
A biliverdin IXβ reductase redox coupling mutation with associated ROS dysregulation has been identified in thrombocytosis cohorts.
Defective BLVRB enzymatic activity involving heme degradation pathway alters metabolic consequences of hematopoietic lineage fate.
Abstract
Human blood cell counts are tightly maintained within narrow physiologic ranges, largely controlled by cytokine-integrated signaling and transcriptional circuits that regulate multilineage hematopoietic specification. Known genetic loci influencing blood cell production account for <10% of platelet and red blood cell variability, and thrombopoietin/cellular myeloproliferative leukemia virus liganding is dispensable for definitive thrombopoiesis, establishing that fundamentally important modifier loci remain unelucidated. In this study, platelet transcriptome sequencing and extended thrombocytosis cohort analyses identified a single loss-of-function mutation (BLVRBS111L) causally associated with clonal and nonclonal disorders of enhanced platelet production. BLVRBS111L encompassed within the substrate/cofactor [α/β dinucleotide NAD(P)H] binding fold is a functionally defective redox coupler using flavin and biliverdin (BV) IXβ tetrapyrrole(s) and results in exaggerated reactive oxygen species accumulation as a putative metabolic signal leading to differential hematopoietic lineage commitment and enhanced thrombopoiesis. These data define the first physiologically relevant function of BLVRB and implicate its activity and/or heme-regulated BV tetrapyrrole(s) in a unique redox-regulated bioenergetic pathway governing terminal megakaryocytopoiesis; these observations also define a mechanistically restricted drug target retaining potential for enhancing human platelet counts.
Introduction
Megakaryocytopoiesis and proplatelet formation represent progressively linked stages of hematopoietic stem cell development that maintain the normal circulating pool of platelets,1 cells critical to normal hemostasis, pathologic thrombosis, and host adaptive immunologic responses.2,3 Platelet generation (∼1 × 1011 cells daily) is largely controlled by the thrombopoietin (TPO)/cellular myeloproliferative leukemia virus (c-MPL) axis, derived by megakaryocyte (MK) commitment form common bipotent MK-erythrocyte progenitors (MEPs).4 Although MKs are reduced in MPL-deficient mice, animals still produce MKs and platelets, implying that hematopoietic stem cells (HSCs) maintain the capacity for lineage fate in the absence of MPL.5 Transcription factors including GATA-1, GATA-2, FOG1/ZFPM1, RUNX1, and NFE2 are important for MK development,2 but none exclusively specify MK fate.1 Whereas human blood counts have a heritable component, known genetic loci account for ∼5% of platelet variability,6,7 highlighting the considerable knowledge gap of genetic pathways regulating physiologic and pathologic thrombopoiesis.
In this study, we applied large-scale platelet transcriptomic sequencing to unmask heme degradation in a novel metabolic pathway controlling megakaryocytopoiesis and platelet production. Whereas nearly 80% of organismal heme is found in circulating erythrocytes, heme degradation is largely studied in the context of reticulendothelial cell-mediated detoxification, with no prior evidence that tetrapyrroles (or their processing enzymes) engage in regulatory functions governing hematopoiesis. Defective oxidation/reductase (redox) activity involving biliverdin IXβ reductase (BLVRB) provides novel insights into the accumulation of reactive oxygen species, with implications for metabolic and bioenergetic determinants of lineage fate. In principle, redox-selected inhibitors represent novel biochemical approaches retaining potential for modulating human platelet counts.
Material and methods
Human subjects and data analyses
All subjects (myeloproliferative neoplasm [MPN; N = 36], reactive thrombocytosis [RT; N = 53], or healthy controls [N = 208]) were enrolled in an institutional review board–approved protocol conducted in accordance with the Declaration of Helsinki,8 applying well-established clinical criteria for phenotypic classification.9,10 Informed consent was obtained from all subjects. Large-scale platelet RNA transcriptomic studies and single nucleotide variant (SNV) identification were completed using the Illumina HiSequation 2000 platform (100 ng RNA/sample); our strategy involved single-end reads and was restricted to nonsynonymous SNVs (nsSNVs) to the exclusion of alternative splicing defects and/or insertion/deletions (in/dels); nsSNVs identified in ≥2 essential thrombocythemia (ET) samples (tier 1) were confirmed by dideoxy sequence analysis prior to expanded genotypic studies, which were completed using Illumina human 610 or 660W single nucleotide polymorphism (SNP) arrays (analyzed using GenomeStudio V2010.2 software). Five distinct genetic models (genotypic, allelic, trend, dominant, and recessive) were applied for the association analyses of each nsSNV (χ2 test, Fisher’s exact test, Cochran-Armigage trend test), comparing different case-control groups with genotypic data available from (1) the 1000 Genomes Project Consortium,11 (2) an internal subset of matched healthy controls, or (3) a cohort subset with RT.8 Statistical comparisons were completed using analysis of variance or Kolmogorov-Smirnov tests, and all statistical analyses were performed using R version 3.1.2.
Hematopoietic, cellular, and biochemical assays
Lentiviruses expressing BLVRB (Lv/BLVRBWT), BLVRBS111L (Lv/BLVRBS111L), or empty virus control (Lv/Control) driven by the cytomegalovirus promoter and containing the puromycin-resistant cassette were generated at the Stony Brook Stem Cell Viral Vector Core and used for transduction of induced pluripotent stem cells (iPSCs) derived from CD34+ human umbilical cords (NCRM1), or human CD34+ hematopoietic stem cell assays as previously described.12 Suspension cultures were maintained in puromycin-selected SFEM II expansion medium supplemented with 50 ng/mL thrombopoietin (MK cultures) or 50 ng/mL thrombopoietin and 2 U/mL erythropoietin (bilineage erythroid/MK expansion).12,13 Cell surface marking and flow cytometry were completed as previously described,12-14 modified for intracellular reactive oxygen species (ROS) accumulation using the cell-permeant fluorogenic probe CellROX Green. Cellular RNA quantitation was performed using fluorescence-based real-time polymerase chain reaction (PCR) technology14 (oligonucleotide primers are provided in supplemental Table 3, available on the Blood Web site).
Specific activity determination of BLVRBWT and BLVRBS111L were completed on bacterially expressed recombinant enzymes at 25°C using flavin mononucleotide or pooled biliverdin (BV) dimethyl esters synthesized by coupled oxidation of heme15 ; RIPA-solubilized cytoplasmic lysates served as the source for cellular BLVRB functional assays,16 and immunodetection was completed using sheep anti-human BLVRB (R&D Systems; 1:100 dilution) and anti-actin monoclonal antibody (mAb) (EMD Millipore; 1:1000).14 Detailed methods are provided in supplemental Methods.
Results
Identification of thrombopoiesis genes using platelet RNASeq
We applied large-scale platelet transcriptome sequencing (RNASeq) to cohorts with ET,8 an MPN subtype causally associated with thrombohemorrhage and characterized by gain-of-function mutations involving Janus kinase 2 (JAK2V617F),17 or CALR in nonmutated JAK2.18,19 To restrict our analyses to candidate genetic defects regulating thrombopoiesis (to the exclusion of platelet functional abnormalities), our strategy included a validation step using subjects with RT, a pathophysiologic subset infrequently associated with thrombohemorrhagic complications (refer to schema; Figure 1A); none of the RT cohort contained CALR or JAK2V617F mutations. RNASeq of highly purified platelets from 7 ET subjects (4 harbored the JAK2V617F mutation and 3 were genotypically normal [G/G]) was completed in parallel using 5 healthy controls, followed by an iterative algorithm to identify nsSNVs as causally plausible candidate genes (Figure 1B). Of the ∼350 000 SNVs, 186 high-quality nsSNVs were identified, of which 33 are qualified as tier 1 based on a stringent filtering step designed to exclude private mutations (supplemental Table 1); 86% of evaluable gene/SNPs had minor allelic frequencies <2%, validating the strategy for identifying rare novel modifier genes using relatively small sample subsets from a clonally expanded hematopoietic disorder. Expectedly, the candidate nsSNV list included JAK2V617F, although neither the initial screen nor targeted visualization of previously described MPN defects20 including MPL or CALR18,19 were identified. This is not unexpected given the relative infrequency of these mutations or the limitations of our SNV search in identifying small in/dels typical of CALR mutations (in our expanded cohort analysis [see “BLVRB gene/SNV is a general thrombocytosis risk allele”], 3 of 9 JAK2V617F-negative patients were subsequently found to be CALR mutated based on sizing assays). Other than JAK2, none of the gene/SNVs have been described as MPN modifiers or overlapped with genetic loci modulating MK/platelet6 or erythroid7 parameters in humans.
Identification of candidate gene/SNVs in the thrombocytosis cohort. (A) Workflow schema detailing phenotypic cohorts that were genetically studied (sample numbers in parentheses; BLVRBS111L [462C→T] mutant number in brackets) and output SNVs at critical validation and genetic association steps. NL, healthy controls. (B) Schema delineating computational pipeline with algorithmic filtering analyses at critical steps of genetic bioinformatics processing. (C) Heat map was generated using expression profiles21 for the candidate gene subset (N = 29) encompassing oligonucleotide probes on the Affymetrix HG_U133AAofAv2 array (scale bar on right). (D) Hematopoietic lineage schema is displayed as a Wilcoxon signed-rank test of the t statistic (−log10 P) calculating the likelihood that the 29 member gene subset is more greatly expressed relative to all other genes expressed in ≥1 time point by lineage (scale bar on right).
Identification of candidate gene/SNVs in the thrombocytosis cohort. (A) Workflow schema detailing phenotypic cohorts that were genetically studied (sample numbers in parentheses; BLVRBS111L [462C→T] mutant number in brackets) and output SNVs at critical validation and genetic association steps. NL, healthy controls. (B) Schema delineating computational pipeline with algorithmic filtering analyses at critical steps of genetic bioinformatics processing. (C) Heat map was generated using expression profiles21 for the candidate gene subset (N = 29) encompassing oligonucleotide probes on the Affymetrix HG_U133AAofAv2 array (scale bar on right). (D) Hematopoietic lineage schema is displayed as a Wilcoxon signed-rank test of the t statistic (−log10 P) calculating the likelihood that the 29 member gene subset is more greatly expressed relative to all other genes expressed in ≥1 time point by lineage (scale bar on right).
We explored the gene expression patterns of the candidate genes using an atlas of 38 distinct hematopoietic cell types21 (supplemental Table 2); 29 of the genes were represented on the Affymetrix gene array (Figure 1C), and candidate transcripts were enriched on average in early-stage MEP and granulocyte/monocyte progenitor cells compared with noncandidate transcripts in the same cell lineage (Figure 1D). These aggregate lineage expression patterns became more restricted in terminally differentiating MKs and erythroblasts (both early and late stages), with less exaggerated (but present) lineage enrichment in myeloid subsets (colony forming unit [CFU]-granulocyte and early-stage neutrophilic metamyelocytes). Limited expression was observed in lymphoid cells (B cells, T cells, and natural killer cells), collectively highlighting that the candidate genes are enriched in all 3 hematopoietic lineages encompassing the clonal evolution of MPNs and underrepresented (or excluded) in lymphoid cells whose genetic composition is generally considered germline in origin.
BLVRB gene/SNV is a general thrombocytosis risk allele
Genotypic studies of the 33-member SNVs using an expanded ET cohort (N = 36)8 followed by statistical association analyses using genotypic frequencies of healthy controls from the 1000 Human Genomes Project11 or an independently genotyped cohort (N = 208), established that 5 SNVs (excluding JAK2V617F) were associated with the ET phenotype (Table 1). Single mutations involving BLVRB (odds ratio [OR], 31.67; P = .006) or KTN1 (OR, 17.32; P = .0018) represented the strongest thrombocytosis risk alleles. The TM7SF3 SNV was a moderate-risk allele (OR, 2.93; P = .005), whereas SNVs involving QSOX1 (OR, 1.96; P = .06) and FAM40B/STRP2 (OR, 3.02; P = .09) only approached marginal statistical significance. The 5 nsSNVs were distributed almost evenly across the JAK2 allelic spectrum, with no statistical evidence by zygosity analysis for any association with JAK2 allelic burden (Figure 2A). To extend these studies, we established the independent (ie, driver) nature of these SNVs by genotyping a new subject cohort with reactive thrombocytosis (N = 53), a nonclonal disorder of exaggerated platelet production due to interleukin-6 (IL-6)–induced thrombopoietin (TPO) release22 ; this approach restricted the gene/SNVs to the subset functioning as modifiers of platelet production independent of JAK2V617F and/or dominant subclones harboring additional molecular abnormalities.20 Only BLVRBS111L retained its significance as a thrombocytosis risk allele in both ET and RT cohorts (Figure 2B), imputing a fundamentally important thrombopoietic function independent of etiology. Although we did not distinguish between somatic and acquired mutation status, its rare minor allelic frequency in the general population (minor allelic frequency ≤0.005; Table 1) is most consistent with a germline predisposition SNV that is phenotypically unmasked during a hematopoietic stimulus.
Characterization of significant SNVs
| Gene . | Chromosome . | SNV* . | Allele . | Mutation . | ET† . | 1000 Genome . | Matched validation cohort . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control† . | P value . | OR . | 95% CI . | Control† . | P value . | OR . | 95% CI . | ||||||
| BLVRB | 19q13.1-q13.2 | rs149698066 | G/A | S111L | 0.042 | 0.0014 | .0006 | 31.7 | [6.3,159.7] | 0.0048 | .02 | 9.0 | [1.5, 54.8] |
| KTN1 | 14q22.1 | rs137964512 | A/G | T232A | 0.045 | 0.0027 | .0018 | 17.3 | [4.2, 70.8] | 0.0072 | .04 | 6.6 | [1.3, 33.2] |
| TM7SF3 | 12q11-q12 | rs10771314 | G/A | P248L | 0.156 | 0.0594 | .0052 | 2.9 | [1.5, 5.9] | 0.1202 | .42 | 1.4 | [0.7, 2.8] |
| QSOX | 1q24 | rs17855475 | G/C | G200A | 0.171 | 0.0955 | .062 | 2.0 | [0.9, 3.8] | 0.1322 | .35 | 1.4 | [0.7 2.7] |
| FAM40B | 7q32.1 | rs61746947 | T/A | V629D | 0.047 | 0.0160 | .091 | 3.0 | [0.9, 10.1] | 0.0433 | .75 | 1.1 | [0.3, 3.8] |
| Gene . | Chromosome . | SNV* . | Allele . | Mutation . | ET† . | 1000 Genome . | Matched validation cohort . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control† . | P value . | OR . | 95% CI . | Control† . | P value . | OR . | 95% CI . | ||||||
| BLVRB | 19q13.1-q13.2 | rs149698066 | G/A | S111L | 0.042 | 0.0014 | .0006 | 31.7 | [6.3,159.7] | 0.0048 | .02 | 9.0 | [1.5, 54.8] |
| KTN1 | 14q22.1 | rs137964512 | A/G | T232A | 0.045 | 0.0027 | .0018 | 17.3 | [4.2, 70.8] | 0.0072 | .04 | 6.6 | [1.3, 33.2] |
| TM7SF3 | 12q11-q12 | rs10771314 | G/A | P248L | 0.156 | 0.0594 | .0052 | 2.9 | [1.5, 5.9] | 0.1202 | .42 | 1.4 | [0.7, 2.8] |
| QSOX | 1q24 | rs17855475 | G/C | G200A | 0.171 | 0.0955 | .062 | 2.0 | [0.9, 3.8] | 0.1322 | .35 | 1.4 | [0.7 2.7] |
| FAM40B | 7q32.1 | rs61746947 | T/A | V629D | 0.047 | 0.0160 | .091 | 3.0 | [0.9, 10.1] | 0.0433 | .75 | 1.1 | [0.3, 3.8] |
From www.ncbi.nlm.nih.gov/SNP.
Minor allelic frequency.
Gene/phenotypic characterization within thrombocytosis cohorts. (A) Distribution of the 5 nsSNVs among the ET cohort by platelet JAK2V617F allelic burden. (B) ORs with confidence intervals (CIs) were calculated by thrombocytosis phenotype (ET, RT) using genotyped validation controls (N = 208); only BLVRBS111L remains a strong risk allele irrespective of thrombocytosis etiology (RT OR = 10.2; CI, 1.96-53.6; P = .005). (C-D) Hemoglobin and platelet counts plotted by JAK2V617F allelic burden (substratified by zygosity for BLVRB 462C→T [S111L] mutation) were fit by linear regressions.
Gene/phenotypic characterization within thrombocytosis cohorts. (A) Distribution of the 5 nsSNVs among the ET cohort by platelet JAK2V617F allelic burden. (B) ORs with confidence intervals (CIs) were calculated by thrombocytosis phenotype (ET, RT) using genotyped validation controls (N = 208); only BLVRBS111L remains a strong risk allele irrespective of thrombocytosis etiology (RT OR = 10.2; CI, 1.96-53.6; P = .005). (C-D) Hemoglobin and platelet counts plotted by JAK2V617F allelic burden (substratified by zygosity for BLVRB 462C→T [S111L] mutation) were fit by linear regressions.
Substratification analysis designed to dissect relative genotypic contributions on blood cell counts confirmed the importance of JAK2V617F allelic burden on hemoglobin concentration (P = .004),23,24 with a modulating effect in the subset coexpressing BLVRBS111L (P = .07; Figure 2C). These results contrasted to those focusing on platelet counts, where JAK2V617F allelic burden displayed minimal effect,23,24 with a trend for exaggerated platelet counts in the BLVRBS111L cohort (Figure 2D), although firm conclusions are limited by the small subset doubly heterozygous for JAK2V617F/BLVRBS111L.
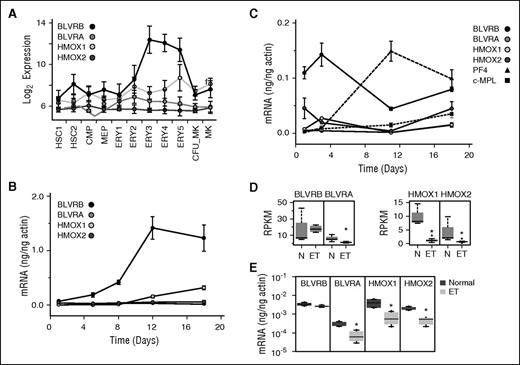
Distinct patterns of BLVRB expression during erythroid/MK speciation
Despite the near-universal presence of hemoproteins across phylogenetic development and their enrichment in erythroid cells (hemoglobin, cytochromes, etc), there is a paucity of information on heme catabolism in either developing or mature hematopoietic cells.25 The BLVRB gene product BV IXβ reductase and its structurally distant homolog BLVRA (BV IXα reductase) function downstream of heme oxygenase(s)-1 (inducible HMOX1) and -2 (constitutive HMOX2) within the heme degradation pathway to catalyze reduction of BV IXα (or IXβ) tetrapyrrole(s) to the potent antioxidants bilirubin (BR) IXα and IXβ.16,26 We studied expression patterns of the 4 heme degradation pathway genes along erythroid/MK (E/Meg) lineage development using data extracted from platelet14 and genetic atlases.21 Initial in silico analyses demonstrated a striking ∼40-fold induction of BLVRB, most pronounced during the terminal phases of erythroid formation (Figure 3A), a pattern that sharply contrasted to that of BLVRA, which remained generally stable (or diminished) throughout the same differentiation period. These patterns were recapitulated in erythropoietin (Epo)-induced CD34+ cultures, again demonstrating exaggerated BLVRB induction that preceded modest induction of HMOX1, with no change in HMOX2 or BLVRA expression (Figure 3B). Comparison of these patterns using Tpo (MK)-directed cultures demonstrated low-level BLVRA, HMOX1, and HMOX2 expression, with essentially no changes in transcript abundance throughout the 18-day culture period (Figure 3C). In contrast, BLVRB maintained the highest transcript abundance throughout the culture, with a predominant peak at an early (day 3/4) time point. Although MK BLVRB peak transcript abundance remained considerably less (ie, ∼10%) than that in late-stage erythroid progenitors, peak levels were comparable to (but preceding) those of the MK commitment marker platelet factor 4 (PF4)14 and remained higher than those of MPL.
Heme degradation pathway characterization during E/Meg development. (A) E/Meg-restricted expression of the heme degradation pathway genes using in silico data.21 A log2 signal intensity <6 is considered background expression. Cell lineage abbreviations are provided in supplemental Table 2. (B-C) Expression patterns of heme degradation pathway genes were quantified by qPCR using human CD34+ cells differentiated along (B) erythroid (Epo) or (C) megakaryocytic (Tpo) lineages; Tpo-directed cultures include Platelet Factor 4 (PF4) and c-MPL transcripts as lineage markers. All results are expressed as actin-normalized means ± standard error of the mean (SEM) from triplicate experiments. (D-E) RNA levels (normalized reads per kilobase of transcript per million reads) of heme degradation pathway genes from (D) ET (N = 7) and normal (N = 5) platelets or as (E) quantified by qPCR (ET, N = 5; normal, N = 5); *P < .05.
Heme degradation pathway characterization during E/Meg development. (A) E/Meg-restricted expression of the heme degradation pathway genes using in silico data.21 A log2 signal intensity <6 is considered background expression. Cell lineage abbreviations are provided in supplemental Table 2. (B-C) Expression patterns of heme degradation pathway genes were quantified by qPCR using human CD34+ cells differentiated along (B) erythroid (Epo) or (C) megakaryocytic (Tpo) lineages; Tpo-directed cultures include Platelet Factor 4 (PF4) and c-MPL transcripts as lineage markers. All results are expressed as actin-normalized means ± standard error of the mean (SEM) from triplicate experiments. (D-E) RNA levels (normalized reads per kilobase of transcript per million reads) of heme degradation pathway genes from (D) ET (N = 7) and normal (N = 5) platelets or as (E) quantified by qPCR (ET, N = 5; normal, N = 5); *P < .05.
These patterns of relative transcript abundance were generally maintained in normal platelets (BLVRB > BLVRA, HMOX1 > HMOX2) as established by extracted reads per kilobase of transcript per million scores and confirmatory quantitative PCR (qPCR; Figure 3D-E). In contrast, ET platelets demonstrated significantly decreased transcript abundance of 3 heme degradation pathway genes (BLVRA, HMOX1, and HMOX2); only the BLVRB transcript level remained comparable to that of healthy controls (vide infra). These aggregate data collectively implicate heme degradation pathway genes in developmentally restricted functions of E/Meg lineage commitment; importantly, the dichotomous platelet expression patterns in ET, coupled with the temporally distinctive BLVRB lineage expression patterns in both erythroid and MK development, collectively suggest a BLVRB function distinct from that of other heme degradation pathway genes.
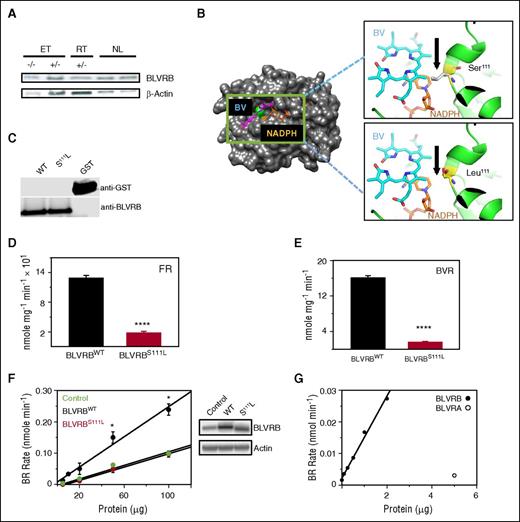
BLVRBS111L is a loss-of-function redox coupler
BLVRB was initially characterized as a flavin reductase27 retaining physiologic relevance primarily as a redox coupler in the presence of methylene blue for treatment of acquired or congenital methemoglobinemia (CYB5A deficiency, Online Mendelian Inheritance in Man #250800). Its function(s) remain additionally enigmatic because it catalyzes formation of the primary rubin generated during fetal hematopoiesis (the IXβ isomer), with no substrate activity toward the predominant BV IXα isomer found in adults.28 BLVRB was readily detectable in healthy control gel-filtered human platelets with no evidence for altered expression in either BLVRBS111L ET or RT platelets (Figure 4A). Comprehensive BLVRB sequence analysis in both RT and ET cohorts identified no additional mutations, prompting more focused study of the BLVRB 462C→T (S111L) heterozygous mutation. Previously characterized crystal structure of the 206 amino acid BV IXβ reductase reveals a monomeric protein with a dinucleotide Rossmann binding fold that preferentially accommodates NAD(P)H as an electron donor with promiscuous binding of various linear tetrapyrroles as electron acceptors27,28 (Figure 4B). Ser111 embedded within the wide D80–K120 substrate binding pocket is structurally homologous to catalytic serines within uridine diphosphate–galactose epimerase (Ser124)29 and ferredoxin–NADP+ reductase (Ser49),30 and uniquely positioned for recognition and/or proton transfer between flavin isoalloxazine and nicotinamide rings.15 Bacterially expressed and purified recombinant BLVRBWT (wild type) and BLVRBS111L demonstrated disparate NAD(P)H-dependent redox coupling using both flavin- and BV-specific substrates (Figure 4C-E), the latter uniquely generated by coupled heme oxidation as verdin-restricted BLVRB activity probes retaining no BLVRA cross-reactivity.15 BLVRBS111L enzymatic activity was defective using flavin mononucleotide (flavin reductase activity; P < .0001) and BV IXβ dimethyl esters (BV reductase [BVR] activity; P < .0001), establishing that the S111L substitution represents a loss-of-function redox mutation with either substrate. These results were confirmed in HEK293 cells using lentivirus(Lv)-induced expression, where enhanced BVR activity was evident in HEK293/BLVRBWT-infected cells with little to no activity in HEK293/BLVRBS111L cells compared with HEK293/Control cells (Figure 4F); residual BVR activity in HEK293/Control and HEK293/BLVRBS111L reflected endogenous BLVRB expression with no evidence for BLVRA cross-reactivity (Figure 4G).
Biliverdin IXβ reductase S111L characterization. (A) BLVRB expression in gel-filtered platelets (10 μg/lane) from representative cohorts that are wild type (−/−) or heterozygous (+/−) for mutant BLVRB462C→T. (B) Schematic globular structure displays BLVRBS111L mutation (green) within the single BV/NAD(P)H binding fold, with higher-resolution ribbon structures (insets) modeled to predict mutant Leu111 or native Ser111 on BV or NADPH interactions; note that Ser111 is uniquely positioned for recognition and/or proton transfer within the Rossmann binding fold, with predicted proximity interference by the hydrophobic Leu111 aliphatic isobutyl side chain. Models were generated using PYMOL software,28 based on NADP/mesobiliverdin IVα ternary (Protein Data Bank ID code 1HE3) and NADP/flavin mononucleotide ternary (Protein Data Bank ID code 1HE4) complexes. (C) Immunoblot of purified recombinant enzymes after thrombin cleavage and glutathione affinity depletion of the GST (glutathione-S-transferase) carrier (50 ng/lane). (D-E) Recombinant BLVRBWT and BLVRBS111L were used for flavin reductase (100 μM flavin mononucleotide) or BVR (20 μM BV dimethyl esters) specific activity determinations15 (N = 6, expressed as mean ± SEM); **** P < .00001. (F) Solubilized lysates from Lv-infected HEK293 cells were used for BVR activity in the presence of 20 μM BV dimethyl esters (N = 3); *P < .05; immunoblot of Lv-infected HEK293 cells (20 μg/lane) is shown. (G) BVR activity assays using recombinant BLVRBWT or commercially available BLVRA.
Biliverdin IXβ reductase S111L characterization. (A) BLVRB expression in gel-filtered platelets (10 μg/lane) from representative cohorts that are wild type (−/−) or heterozygous (+/−) for mutant BLVRB462C→T. (B) Schematic globular structure displays BLVRBS111L mutation (green) within the single BV/NAD(P)H binding fold, with higher-resolution ribbon structures (insets) modeled to predict mutant Leu111 or native Ser111 on BV or NADPH interactions; note that Ser111 is uniquely positioned for recognition and/or proton transfer within the Rossmann binding fold, with predicted proximity interference by the hydrophobic Leu111 aliphatic isobutyl side chain. Models were generated using PYMOL software,28 based on NADP/mesobiliverdin IVα ternary (Protein Data Bank ID code 1HE3) and NADP/flavin mononucleotide ternary (Protein Data Bank ID code 1HE4) complexes. (C) Immunoblot of purified recombinant enzymes after thrombin cleavage and glutathione affinity depletion of the GST (glutathione-S-transferase) carrier (50 ng/lane). (D-E) Recombinant BLVRBWT and BLVRBS111L were used for flavin reductase (100 μM flavin mononucleotide) or BVR (20 μM BV dimethyl esters) specific activity determinations15 (N = 6, expressed as mean ± SEM); **** P < .00001. (F) Solubilized lysates from Lv-infected HEK293 cells were used for BVR activity in the presence of 20 μM BV dimethyl esters (N = 3); *P < .05; immunoblot of Lv-infected HEK293 cells (20 μg/lane) is shown. (G) BVR activity assays using recombinant BLVRBWT or commercially available BLVRA.
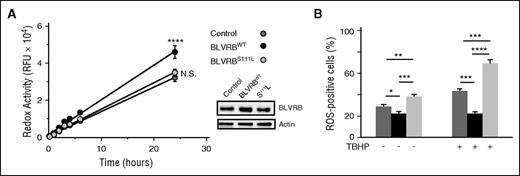
BLVRB redox function alters cellular ROS accumulation
Previous data have suggested that a BV/BR redox cycle provides a mechanism for neutralization of ROS and cytoprotection.16,17 Accordingly, we hypothesized that defective BLVRBS111L redox coupling could affect ROS accumulation, a requisite upstream signaling messenger of MK differentiation,31,32 and stem cell quiescence during migration from hypoxic (low ROS) osteoblastic to oxygen-rich (high ROS) vascular niches.33,34 Hematopoietic-derived (CD34+) iPSCs infected with individual lentiviruses established that iPSC/BLVRBWT cells (expressing BLVRB approximately twofold greater than control) retained enhanced redox activity (P = .001) compared with both control and iPSC/BLVRBS111L cells; redox coupling in iPSC/BLVRBS111L paralleled that of control iPSCs (Figure 5A). Enhanced iPSC/BLVRBWT-associated redox coupling was associated with statistically lower baseline ROS accumulation, whereas baseline ROS accumulation was highest in iPSC/BLVRBS111L cells (Figure 5B). Incubation with the organic peroxide tert-butyl hydroperoxide (TBHP) as the oxidant stress source protected against ROS accumulation in iPSC/BLVRBWT cells using a TBHP concentration causing ROS accumulation in ∼50% of either control or iPSC/BLVRBS111L cells (EC50 = 200 μM). The ROS-neutralizing characteristics of iPSC/BLVRBWT cells contrasted sharply to those of iPSC/BLVRBS111L, which demonstrated exaggerated TBHP-induced ROS accumulation when compared with either iPSC/BLVRB (P < .00001) or control iPSCs (P < .0002). Thus, the BLVRB-mediated ROS-neutralizing effect appears comparable to that of BLVRA,16,35 whereas defective BLVRBS111L activity enhances ROS at baseline with exaggerated ROS accumulation on oxidant stress. Importantly, as heterozygous models designed to phenocopy zygosity state in thrombocytosis cohorts, these collective data establish a BLVRBS111L dominant inhibitory effect in cells expressing both native and mutant BLVRB.
Differential patterns of ROS handling in genetically modified CD34+ iPS cells. (A) Genetically modified CD34+ NCRM1 iPSCs (1 × 104 cells per well) were loaded with 0.1 (v/v) resazurin for time-dependent spectrofluorimetric detection (530-nm excitation, 590-nm emission) of reduced resorufin as a quantitation of cellular redox coupling; data expressed as mean ± SEM of relative fluorescent units (N = 6); cellular BLVRB expression is shown, 20 μg lysates/lane; ****P < .0001. (B) Genetically modified NCRM1 iPSCs (1 × 105/mL) were treated (or not) with 200 μM TBHP for 1 hour at 37°C, followed by flow cytometric quantification of ROS-expressing cells after a 60-minute loading with cell-permeable 500 nM CellROX green as indicator (N = 6); *P < .05; **P < .01; ***P < .001; ****P < .0001.
Differential patterns of ROS handling in genetically modified CD34+ iPS cells. (A) Genetically modified CD34+ NCRM1 iPSCs (1 × 104 cells per well) were loaded with 0.1 (v/v) resazurin for time-dependent spectrofluorimetric detection (530-nm excitation, 590-nm emission) of reduced resorufin as a quantitation of cellular redox coupling; data expressed as mean ± SEM of relative fluorescent units (N = 6); cellular BLVRB expression is shown, 20 μg lysates/lane; ****P < .0001. (B) Genetically modified NCRM1 iPSCs (1 × 105/mL) were treated (or not) with 200 μM TBHP for 1 hour at 37°C, followed by flow cytometric quantification of ROS-expressing cells after a 60-minute loading with cell-permeable 500 nM CellROX green as indicator (N = 6); *P < .05; **P < .01; ***P < .001; ****P < .0001.
BLVRBS111L differential effects on hematopoietic lineage commitment
Potential consequences of differential BLVRB redox coupling and ROS handling on lineage commitment were evaluated using primary CD34+ HSCs transduced with Lv/BLVRBWT, Lv/BLVRBS111L, and Lv/Control. Suspension cultures prior to terminal differentiation (day 0) demonstrated no differences in cellular viability (Figure 6A), excluding potential ROS-damaging effects in CD34+/BLVRBS111L cells that contained >70% mutant T alleles as established by pyrosequencing. Furthermore, there was general expansion of CD34+/BLVRBWT and CD34+/BLVRBS111L progenitor cells compared with CD34+/Control, with results that were evident both by cell number and by flow cytometric quantification of gated, live (7-amino-actinomycin D [7-AAD] negative) CD34+ cells (Figure 6B). Comparative effects of BLVRB-associated redox coupling/ROS handling on proliferative potential and lineage commitment were established using primary CD34+ methylcellulose multipotential progenitor cultures, results that confirmed exaggerated aggregate colony formation in Lv/BLVRBWT- and Lv/BLVRBS111L-transduced cells compared with Lv/Control (Figure 6C). Conversely, there was disproportionate expansion of primitive CFU-granulocytes/erythrocytes/monocytes/MKs in CD34+/BLVRBS111L cells (P = .001), and an absolute increase of burst forming units, erythroid colonies in CD34+/BLVRBWT cells (P = .001). These results confirmed that the proliferative effect occurred independently from an effect on lineage commitment. Collagen-based cultures designed to specifically quantify MK progenitor potential at a single-cell level demonstrated a statistically significant increase of CD41+ CFU-MKs in CD34+/BLVRBS111L cells (P < .01) with no increased CFU-MKs in CD34+/BLVRBWT cells (Figure 6D), confirming disparate effects on MK lineage commitment and a preserved proliferative function distinct from its redox capacity.
BLVRBS111L hematopoietic effects on ROS accumulation and lineage fate. (A-B) CD34+ HSCs transduced with Lv/BLVRBWT, Lv/BLVRBS111L, or Lv/Control were puromycin-selected and expanded for 48 hours prior to terminal differentiation (day 0). Genetically modified cells were assessed for viability using (A) trypan blue exclusion (N = 4) or (B) cell quantification and flow cytometry. Cell quantification (N = 6) is expressed as relative fold-increase to control, and flow cytometric quantification was determined using gated, live (7-AAD negative) CD34+ cells (N = 4); flow cytometry gate (forward scatter [FSC], size vs side scatter [SSC], side scatter as a measure of complexity) used for quantification of 7-AAD negative CD34+ cells and for ROS tracking is shown. (C-D) Genetically modified CD34+ HSCs were puromycin selected and plated for determination of lineage fate using (C) multipotential progenitor assays (N = 4 experiments) or (D) CFU-MK determinations (N = 3 experiments). (E) Representative flow cytometric analysis of developing MKs dually labeled for ROS accumulation and CD41 at discrete time points (LV/control-infected CD34+ HSCs are shown). (F) Cumulative distribution plots across ROShigh subsets generated by flow cytometric quantification of MK phenotypic parameters (ROS and CD41 [Tpo cultures], or CD41 [CD41+/GlyA− [bilineage Tpo/Epo cultures]) are displayed by culture conditions and time points for genetically-modified CD34+/BLVRBWT, CD34+/BLVRBS111L, and CD34+/Control; results are from a single representative experiment repeated on 3 occasions; P values for pairwise comparisons were calculated over 200 bootstrapped samples. (G) Percentage of CD41−/GlyA+ erythroid cells in day 10 bilineage cultures (N = 3). (H) UV-visible spectroscopy of day 10 bilineage cultures (normalized to the peak Soret absorbance of oxyhemoglobin [λ414]) demonstrates no differences in methemoglobin (λ630) accumulation across the genotypes. For all panels except F, results are expressed as mean ± SEM; *P < .05, **P < .01, ***P < .001, N.S., not significant.
BLVRBS111L hematopoietic effects on ROS accumulation and lineage fate. (A-B) CD34+ HSCs transduced with Lv/BLVRBWT, Lv/BLVRBS111L, or Lv/Control were puromycin-selected and expanded for 48 hours prior to terminal differentiation (day 0). Genetically modified cells were assessed for viability using (A) trypan blue exclusion (N = 4) or (B) cell quantification and flow cytometry. Cell quantification (N = 6) is expressed as relative fold-increase to control, and flow cytometric quantification was determined using gated, live (7-AAD negative) CD34+ cells (N = 4); flow cytometry gate (forward scatter [FSC], size vs side scatter [SSC], side scatter as a measure of complexity) used for quantification of 7-AAD negative CD34+ cells and for ROS tracking is shown. (C-D) Genetically modified CD34+ HSCs were puromycin selected and plated for determination of lineage fate using (C) multipotential progenitor assays (N = 4 experiments) or (D) CFU-MK determinations (N = 3 experiments). (E) Representative flow cytometric analysis of developing MKs dually labeled for ROS accumulation and CD41 at discrete time points (LV/control-infected CD34+ HSCs are shown). (F) Cumulative distribution plots across ROShigh subsets generated by flow cytometric quantification of MK phenotypic parameters (ROS and CD41 [Tpo cultures], or CD41 [CD41+/GlyA− [bilineage Tpo/Epo cultures]) are displayed by culture conditions and time points for genetically-modified CD34+/BLVRBWT, CD34+/BLVRBS111L, and CD34+/Control; results are from a single representative experiment repeated on 3 occasions; P values for pairwise comparisons were calculated over 200 bootstrapped samples. (G) Percentage of CD41−/GlyA+ erythroid cells in day 10 bilineage cultures (N = 3). (H) UV-visible spectroscopy of day 10 bilineage cultures (normalized to the peak Soret absorbance of oxyhemoglobin [λ414]) demonstrates no differences in methemoglobin (λ630) accumulation across the genotypes. For all panels except F, results are expressed as mean ± SEM; *P < .05, **P < .01, ***P < .001, N.S., not significant.
The Tpo differentiation suspension culture across the genotypes confirmed that >98% of live cells were ROShigh and CD41+ (αIIBβ3) acquisition occurred almost exclusively within ROShigh subsets (Figure 6E). These data are consistent with a requisite developmental ROS signal during terminal megakaryocytopoiesis25,26 and provided a surrogate marker for BLVRB redox activity that remained below threshold sensitivity throughout the 10-day culture period.36 We tracked the distribution of ROS accumulation within ROShigh subsets and noted ROS-attenuating effects of CD34+/BLVRBWT that contrasted with exaggerated ROS accumulation of CD34+/BLVRBS111L; these differences were most pronounced at day 4, but were still evident at day 10 of terminal differentiation (P < .05; Figure 6F). Exaggerated ROS accumulation in MK/BLVRBS111L cells at Day 4 corresponded to temporally earlier CD41 induction (P < .05); in contrast, ROS-attenuating effects of day 4 MK/BLVRBWT cells were not associated with diminished CD41 acquisition at day 4, likely explained by a threshold ROS accumulation sufficient to maintain its “priming” effect on MK development.33 Bilineage (Tpo/Epo) culture conditions designed to address putative effects on MEP balance12,13 demonstrated no differences among the genotypes on MK partitioning (CD41+/glycophorin A− [GlyA−] cells), suggesting that BLVRBS111L MK effect was restricted to postcommitment expansion and not MEP partitioning; interestingly, there was a general expansion in both BLVRBWT (P = .03) and BLVRBS111L (P = .06) erythroid (CD41−/GlyA+) compartments compared with Lv/Control (Figure 6G). Finally, we completed UV-visible spectroscopy of late-stage bilineage cultures (Figure 6H) (in which CD41−/GlyA+ erythroid fraction accounts for >95% of cells13 ), with no evidence for methemoglobin accumulation in BLVRBS111L erythroid cells.27,37
Discussion
Our data provide the first evidence linking redox activity within the heme degradation pathway to MK lineage fate and physiologically relevant platelet expansion in humans. Support for this conclusion stems from (1) genetic association studies in both clonal and nonclonal disorders of thrombocytosis, (2) biochemical analyses using complementary NADPH-dependent enzymatic determinations of both flavin and BV substrates, (3) documentation that defective redox activity is associated with ROS mishandling as a metabolic cellular consequence in both iPSCs and primary hematopoietic cells, and (4) evidence for disparate effects on MK lineage fate in suspension and collagen-based culture systems. Although BLVRBS111L is clearly associated with ROS mishandling as a putative explanation for accelerated differentiation (or “priming”)33 during a developmentally regulated stage of megakaryocytopoiesis, a direct metabolic effect related to tetrapyrrole coupling (unrelated to the surrogate ROS accumulation) remains plausible and is currently under investigation; similarly, we cannot exclude an additive effect of enhanced ROS-associated proplatelet formation during late-stage MK development.38
Interestingly, there is minimal evidence implicating any of the heme degradation pathway components in hematopoietic development, which is an unexpected finding in our study. Despite evidence for cytoprotective effects of HMOX and BLVRB,16,39 heme degradation is generally considered a catabolic pathway, predominantly active in cells of the reticulo-endothelial system to process pro-oxidant (free) heme generated during erythroid senescence or turnover of cytochrome p450 enzymes. Within the subset of heme degradation pathway genes, BLVRB expression (to the exclusion of the other 3 genes HMOX1, HMOX2, and BLVRA) demonstrated a striking induction during terminal erythropoiesis and a restricted temporal pattern of expression during megakaryocytopoiesis. Thus, our data are consistent with a recent report highlighting the importance of HMOX1 in murine erythroid development25 and extend this observation to a lineage-restricted effect on megakaryocytopoiesis. Importantly, the comparatively disparate expression patterns also suggest that BLVRB maintains functional effects distinct from those of other heme degradation components.
To date, it remains unestablished if BLVRB megakaryocytic effects are restricted to its activity as a flavin reductase, in partnered electron exchange with an unidentified protein, or as a verdin-regulated redox coupler. The latter mechanism implies a developmentally coordinated hematopoietic function(s) regulated by heme degradation and isomer-restricted BV IXβ (or IXδ, IXγ) generation. Indeed, this conclusion seems paradoxical given our current understanding of BV/BR isomeric patterns in adults. All natural BVs are formed by ring cleavage of preassembled heme and not by de novo assembly from smaller units40 ; although cleavage could occur at any of the 4 meso bridge carbons, there is striking regioselectivity of HMOX for the α-meso (IXα) carbon bridge with limited generation of BLVRB-specific IXβ, IXδ, and IXγ isomers.28 Interestingly, BV isomer generation appears distinct in the fetus where BV IXβ may predominate,41 although the mechanism for BV IXβ generation remains enigmatic. Nonetheless, BLVRB binds promiscuously (and nonproductively) to other tetrapyrroles such as protohemin27 or BV IXα,28 suggesting that its redox activity could be modulated by endogenous cellular heme byproducts with resultant effects on redox coupling and lineage commitment (ie, function as a metabolically regulated cellular rheostat34 ).
The loss-of-function redox mutation causes defective ROS handling in the background of endogenous BLVRB expression, establishing a dominant inhibitory mechanism of action. This is somewhat unexpected because the crystal structure provides no convincing evidence for dimerization sequences28 (either homo- or heterodimers) as one putative explanation, although an alternative model of enzymatically locked substrate (and/or cofactor) binding remains plausible. Indeed, a compulsory ordered kinetic mechanism has been proposed in which NADPH binding followed by substrate results in sequential release of product and oxidized cofactor.28 A definitive explanation will require crystallization and structure determination of BLVRBS111L; indeed, this issue may be relevant for previous suggestions imputing the existence of a BV/BR redox cycle and progressively amplified BR antioxidant functions.16 Finally, the dominant inhibitory mechanism predicts that low-level BLVRB 462C→T allelic expression could have exaggerated cellular effects in the background of wild-type C alleles.
Low intracellular ROS levels evident in quiescent HSCs are regulated by the FoxO family of transcription factors,42 and the primary non-mitochondrial sources for ROS production are NADPH oxidases. In addition, the thiol balance also contributes to hematopoietic progenitor cell mobilization, as cellular redox status can be regulated by free or protein-incorporated thiols. Previous data have established that p22hox-dependent NADPH oxidase activity is responsible for ROS production and required for MK differentiation.32 Despite the importance of ROS signaling in cell quiescence and lineage fate,33 there are no identified ROS-regulating mutations that modulate human platelet counts, and we would speculate that the temporally restricted nature of BLVRB-associated ROS priming provides the developmentally regulated signal for accelerated MK differentiation. Using a bilineage model of erythro/megakaryocytopoiesis, we saw no evidence for MK lineage partitioning, most consistent with a model of postcommitment megakaryocytopoiesis and not altered E/Meg lineage balance.13 Nonetheless, it is intriguing that our limited cohort analysis demonstrated reciprocal effects on hemoglobin and platelet counts in BLVRBS111L subsets, observations that need to be confirmed in expanded MPN subtypes including polycythemia vera. Expectedly, we saw no evidence that BLVRBS111L caused methemoglobinemia in the setting of functionally intact cytochrome b5 reductase,27 although statistically significant erythroid expansion was evident in BLVRBWT multipotential progenitor and bilineage cultures. The latter observation remains under investigation but suggests an erythroid function in redox-regulated bioenergetic metabolism, with possible effects on stage-specific erythropoiesis.43,44
BLVRB (and the BLVRA homolog) are unusual, possibly unique, in their ability to use either NADPH or NADH as cofactors with a different pH optimum for each cofactor.45 Nonetheless, BLVRB accommodates NADPH more favorably,28 and cofactor binding is required for apoprotein stability; furthermore, broad substrate discrimination occurs by steric constraints for chemical groups adjacent to the electrophilic C10 methene carbon. When integrated with our data, this model suggests that development of BLVRB-selective redox inhibitors with thermodynamically distinct chemical modifications represents logical approaches to selectively alter a regulatory pathway controlling MK lineage expansion and human platelet counts.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank M. Golightly (Stony Brook University) for assistance with flow cytometric analyses, I. Carrico and E. Boon (Stony Brook University) for assistance with heme oxidation studies, M. Seeliger and R. Rizzo (Stony Brook University) for assistance with computational modeling, and P. Pereira and S. Macedo-Ribeiro (University of Porto) for helpful discussions.
This work was supported by National Institutes of Health (NIH)/National Heart Lung, and Blood Institute (NHLBI) grant HL091939, NIH/NHLBI DNA Resequencing and Genotyping Service (subcontract G175), the New York State Stem Cell Foundation (grant C026716), and the Burroughs Wellcome Fund (W.F.B.); grants from the Stony Brook Targeted Research Opportunities (S.W.); and NIH/NHLBI grant HL12945 (N.M.N.).
Authorship
Contribution: W.F.B. and S.W. conceptualized the study and formulated the hypothesis; W.F.B. and S.W. designed the research; S.W., Z.L, D.V.G., B.Z., L.Z., L.E.M., N.M., T.J.M., N.M.N., and W.F.B. performed research; W.F.B. and S.W. wrote the paper; and W.F.B. directed the overall research.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Wadie F. Bahou, Department of Medicine and the Stony Brook Stem Cell Facility, Health Sciences Center T15-040, Stony Brook University School of Medicine, Stony Brook, NY 11794-8151; e-mail: wadie.bahou@sbumed.org.

![Figure 1. Identification of candidate gene/SNVs in the thrombocytosis cohort. (A) Workflow schema detailing phenotypic cohorts that were genetically studied (sample numbers in parentheses; BLVRBS111L [462C→T] mutant number in brackets) and output SNVs at critical validation and genetic association steps. NL, healthy controls. (B) Schema delineating computational pipeline with algorithmic filtering analyses at critical steps of genetic bioinformatics processing. (C) Heat map was generated using expression profiles21 for the candidate gene subset (N = 29) encompassing oligonucleotide probes on the Affymetrix HG_U133AAofAv2 array (scale bar on right). (D) Hematopoietic lineage schema is displayed as a Wilcoxon signed-rank test of the t statistic (−log10 P) calculating the likelihood that the 29 member gene subset is more greatly expressed relative to all other genes expressed in ≥1 time point by lineage (scale bar on right).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/128/5/10.1182_blood-2016-02-696997/4/m_699f1.jpeg?Expires=1769175936&Signature=U5PIZmfApsdXvU61fcEoOkQfMlw75pTCsu0m7EquVRYKrjVARwkGye122HWb37VxBZNXSJZuKdA39rg4lCYvTCqzJy5qHcRg6mgzTiUIszIzJ6HHc85rq12hF0ohWqbCQUbva5DU3Mp73mwoOtRi94rrkhSp6~ajrOgLWp0bfp1uzZCn9CQHuBbMp4tJee-JAqDGaSQS4nx9rXkhkV8tXJYzCgGNl3ArvD~ou~BRQedaD~D9Xz2gMM~ntoVwMtM4tp50kyCAJ-C18PETvuJLgcbQgOew38YQm-Tvp1nVdznVJBb3898pIXi33yEkTIHVTNM31l-kVsflamNHoS2dow__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 2. Gene/phenotypic characterization within thrombocytosis cohorts. (A) Distribution of the 5 nsSNVs among the ET cohort by platelet JAK2V617F allelic burden. (B) ORs with confidence intervals (CIs) were calculated by thrombocytosis phenotype (ET, RT) using genotyped validation controls (N = 208); only BLVRBS111L remains a strong risk allele irrespective of thrombocytosis etiology (RT OR = 10.2; CI, 1.96-53.6; P = .005). (C-D) Hemoglobin and platelet counts plotted by JAK2V617F allelic burden (substratified by zygosity for BLVRB 462C→T [S111L] mutation) were fit by linear regressions.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/128/5/10.1182_blood-2016-02-696997/4/m_699f2.jpeg?Expires=1769175936&Signature=XUXRTOPSV~U7Wiyzd7GpL7AuUkfrrgPMgl6HsaY4shxYmFbVH4gLob-eD3eEWPftOofPx6NunoLXOa-eHV~F01R7M-7hm2oxN-MWCpX8lrQk2NEXXhwSxdVnnHxLmJ0jssRpcF42l6M6-IeImjD0nWTAShTVRqd0UZDkzK9Mo02ddRU3k1O7g7fEEJC5~ZJ1Nkx~NQGi9zUdqpeflugy6kbW8MmfasvSAY6RdfH1vOE55Hyf-qO4tSulGNfFvuNNzd35okFyW8BTiDC3C5QksViK9iQribkPrV2GeJSM3vX3k1mYNJRMTsGyAzPzr0FZD7t4FCuKgwXGMZfXtsbUVg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 6. BLVRBS111L hematopoietic effects on ROS accumulation and lineage fate. (A-B) CD34+ HSCs transduced with Lv/BLVRBWT, Lv/BLVRBS111L, or Lv/Control were puromycin-selected and expanded for 48 hours prior to terminal differentiation (day 0). Genetically modified cells were assessed for viability using (A) trypan blue exclusion (N = 4) or (B) cell quantification and flow cytometry. Cell quantification (N = 6) is expressed as relative fold-increase to control, and flow cytometric quantification was determined using gated, live (7-AAD negative) CD34+ cells (N = 4); flow cytometry gate (forward scatter [FSC], size vs side scatter [SSC], side scatter as a measure of complexity) used for quantification of 7-AAD negative CD34+ cells and for ROS tracking is shown. (C-D) Genetically modified CD34+ HSCs were puromycin selected and plated for determination of lineage fate using (C) multipotential progenitor assays (N = 4 experiments) or (D) CFU-MK determinations (N = 3 experiments). (E) Representative flow cytometric analysis of developing MKs dually labeled for ROS accumulation and CD41 at discrete time points (LV/control-infected CD34+ HSCs are shown). (F) Cumulative distribution plots across ROShigh subsets generated by flow cytometric quantification of MK phenotypic parameters (ROS and CD41 [Tpo cultures], or CD41 [CD41+/GlyA− [bilineage Tpo/Epo cultures]) are displayed by culture conditions and time points for genetically-modified CD34+/BLVRBWT, CD34+/BLVRBS111L, and CD34+/Control; results are from a single representative experiment repeated on 3 occasions; P values for pairwise comparisons were calculated over 200 bootstrapped samples. (G) Percentage of CD41−/GlyA+ erythroid cells in day 10 bilineage cultures (N = 3). (H) UV-visible spectroscopy of day 10 bilineage cultures (normalized to the peak Soret absorbance of oxyhemoglobin [λ414]) demonstrates no differences in methemoglobin (λ630) accumulation across the genotypes. For all panels except F, results are expressed as mean ± SEM; *P < .05, **P < .01, ***P < .001, N.S., not significant.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/128/5/10.1182_blood-2016-02-696997/4/m_699f6.jpeg?Expires=1769175936&Signature=Yb7bD0CccCPCoenMtxWn6gMgOSAVDzaNXZCvGlWkm6JcZaVBMXuzANp7DwWr5ympsojh~AnHGJ9E4z0KFqrzGXmOMj5eOV~HAnItSwiUsNMQ2yMyRBI0nwu7HwQcIIdqdtnTuLeQoJPMde68zcY6UJUicUdnnLy2A~s4yOC0l1iV1Ghns0oCkf3Es1ysUtQto-tWSZuI7jUUFBZSxBPOqEsOrDhBW19fGdDfqRbPPoPFX75eg~EjOdTzYiWdMJIeHZf-EswofqqXmZVWLtTKzSVyPjlxiZD0YcqLVj~ST9mM7oZWp8rHmJpYUwObcZQrp1ktUgagoeqYA3J3WjYtFg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal