Key Points
CD40/IL-4–stimulated CLL cells release EVs enriched with specific miRNAs including miR-363.
Transfer of CLL-EVs to autologous CD4+ T cells enhances migration and immune synapse formation interactions with tumor cells.
Abstract
The complex interplay between cancer cells, stromal cells, and immune cells in the tumor microenvironment (TME) regulates tumorigenesis and provides emerging targets for immunotherapies. Crosstalk between CD4+ T cells and proliferating chronic lymphocytic leukemia (CLL) tumor B cells occurs within lymphoid tissue pseudofollicles, and investigating these interactions is essential to understand both disease pathogenesis and the effects of immunotherapy. Tumor-derived extracellular vesicle (EV) shedding is emerging as an important mode of intercellular communication in the TME. In order to characterize tumor EVs released in response to T-cell–derived TME signals, we performed microRNA (miRNA [miR]) profiling of EVs released from CLL cells stimulated with CD40 and interleukin-4 (IL-4). Our results reveal an enrichment of specific cellular miRNAs including miR-363 within EVs derived from CD40/IL-4–stimulated CLL cells compared with parental cell miRNA content and control EVs from unstimulated CLL cells. We demonstrate that autologous patient CD4+ T cells internalize CLL-EVs containing miR-363 that targets the immunomodulatory molecule CD69. We further reveal that autologous CD4+ T cells that are exposed to EVs from CD40/IL-4–stimulated CLL cells exhibit enhanced migration, immunological synapse signaling, and interactions with tumor cells. Knockdown of miR-363 in CLL cells prior to CD40/IL-4 stimulation prevented the ability of CLL-EVs to induce increased synapse signaling and confer altered functional properties to CD4+ T cells. Taken together, these data reveal a novel role for CLL-EVs in modifying T-cell function that highlights unanticipated complexity of intercellular communication that may have implications for bidirectional CD4+ T-cell:tumor interactions within the TME.
Introduction
Chronic lymphocytic leukemia (CLL) is one of the most common adult B-cell malignancies and is characterized by the clonal expansion of CD5+ mature B cells in lymph nodes and the peripheral blood.1,2 Emerging research suggests that the complex interplay between leukemic cells and stromal cells, including immune cells, in the lymphoid tissue tumor microenvironment (TME) critically regulates tumor progression and the licensing of immune evasion.3-6
CLL tumor cells have been shown to evade proximal antitumor T-cell activity by delivery of inhibitory signals (including via the PD-L1:PD-1 immune checkpoint pathway)6 into T cells to downregulate lytic immune synapse activity. However, proliferating CLL cells colocalize with CD4+ T cells within lymph node pseudofollicles (proliferation centers), suggesting intercellular activatory signaling and tumor-promoting interactions.7 Understanding tumor T-cell crosstalk will be essential to overcome complex protumorigenic immune-regulatory mechanisms in the TME. Moreover, biological insight into tumor:CD4+ T-cell communication will help understand the effect of activating T cells in the CLL TME using novel immune checkpoint immunotherapy such as anti-PD-1 antibody drugs.8-10
Although cell-to-cell contact (ligand:receptor interactions) and exchange of soluble factors (classical paracrine growth factor signaling loops) are key mediators of intercellular communication within the TME, more recently, extracellular vesicle (EV) shedding has emerged as another mode of cell-cell signaling.11 EVs comprise exosomes (30-100 nm), which have an endocytic origin and larger microvesicles, which are released from the plasma membrane. Categorizing vesicles based on their size or subcellular origin remains problematic,12 and whether or not 1 type of vesicle is biologically more relevant than another is currently unclear within the field.13
Transfer of EVs and their cargo, which includes proteins, lipids, messenger RNA (mRNA), and microRNAs (miRNAs [miRs]), from cancer cells to other TME cell types has been the subject of intense study in the solid cancers13 but has been relatively understudied in lymphoid malignancies. Recent work has revealed that B-cell receptor (BCR) activation, a critical survival pathway within the CLL TME, enhances secretion of CLL exosomes that carry a distinct miRNA signature compared with healthy donors.14 Moreover, CLL-EVs and their molecular cargo are actively transferred to stromal cells inducing intracellular signaling and the reprogramming of these cells into proinflammatory cancer-associated fibroblasts.5,15,16
The release of EVs is enhanced when primary murine B cells are stimulated by T-cell signals17 but the effects of CD40 and IL-4 stimulation, among the most important immune-derived TME stimuli,18,19 on the production and content of CLL-EVs has not previously been investigated. Here, we determine those miRNA selectively enriched or depleted in CLL-EVs following CD40/interleukin-4 (IL-4) stimulation (CD40/IL-4 CLL-EVs). We show that miR-363 is highly enriched in CD40/IL-4 CLL-EVs and that a principal target for this miRNA is the T-cell immunoregulatory receptor CD69. Critically, we show that the transfer of CLL-EVs into autologous CD4+ T cells increases motility rate, immunological synapse signaling, and interactions with tumor cells. These results define a novel immunomodulatory mechanism in CLL induced by EVs and their molecule cargo, which might have important implications for crosstalk between the TME and CLL cells.
Materials and methods
Patient samples
Mononuclear cells were isolated from heparinized whole blood from CLL patients (local research ethics committee approval with informed consent), using Histopaque density gradient media (Sigma-Aldrich) with density gradient centrifugation. CD19+CD5+ CLL were purified using negative selection for the EV isolation studies (MACS system; Miltenyi Biotec). Patient characteristics are presented in supplemental Table 1 (available on the Blood Web site).
EV isolation
CD19+ CLL cells were cultured in Aim V with AlbuMAX serum-free media (Invitrogen) at a concentration of 3 × 106/mL. To stimulate the release of CLL B-cell EVs, CLL cells were incubated with recombinant human CD40L (sCD154) (1 µg/mL; R&D Systems) and IL-4 (20 ng/mL; R&D Systems) and anti-CD40 antibody (clone EA-5) at 1 µg/mL (sc65264; Santa Cruz Biotechnology) for 36 hours. CLL-EVs were harvested from 36 × 106 cultured leukemic cells at 4°C by differential centrifugation of culture supernatant. Supernatants were spun at 250g for 5 minutes followed by 2000g for 10 minutes to pellet cells and debris, and 10 000g for 30 minutes to pellet debris and larger membrane-bound particles. EVs were pelleted by centrifugation at 100 000g for 110 minutes, washed in phosphate-buffered saline then repelleted, and finally resuspended in culture media for analysis (see supplemental Methods for details).14
miRNA microarrays
miRNA extracts were made using a miRvana miRNA isolation kit (Applied Biosystems). RNA concentration was measured (NanoDrop 1000 Spectrophotometer; Thermo Fisher Scientific) and equal amounts were reverse transcribed using MegaPlex RT primers and a TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems). Equal volumes were loaded on human microRNA array (A) plates (containing assays for 377 miRNA; Applied Biosystems) and reactions carried out on a 7900HT Real Time PCR system.20 Forty polymerase chain reaction (PCR) cycles were carried out and cycle threshold (Ct) values >35 were not included in the analysis. All Ct values were normalized to the global plate mean of expressed miRNA Ct values, and ΔCt values were calculated. In order that each plate had approximately the same mean Ct value, both cellular and extracellular vesicle samples loaded on the same plate.
CLL cell transfection
CLL cells were cultured in the presence or absence of CD40/IL-4 stimulation in 6-well plates at 1 × 107/mL with a volume of 2 mL per well. Either miRCURY LNA Power microRNA inhibitor miR-363 (Exiqon) or miRCURY inhibitor negative control A (Exiqon) were added to the cultures at a final concentration of 50 nM for 48 hours before harvesting. In order to knockdown Rab27A and Rab27B, antisense LNA GapmeRs (Exiqon) were used at 100 nM for 96 hours. Negative control antisense LNA GapmeR (Exiqon) was also used. Cells were harvested and degree of knockdown checked by western with anti-Rab27 antibody (Proteintech) at 1:1000.
Immune synapse assay
Motility video microscopy
The motility studies were performed and analyzed as previously described (see supplemental Methods for details).22
Statistical methods
Statistical methods used in this study are outlined within the respective methodology sections (see supplemental Methods for details) and figure legends. Data were compared with the use of either the 2-tailed Student t test or the nonparametric Mann-Whitney test. Analysis was done using Prism version 5 software. P < .05 was considered statistically significant.
Results
Characterization and miRNA profiling of CLL-EVs derived from CD40/IL-4–stimulated CLL cells reveals enrichment of specific miRNA content including miR-363
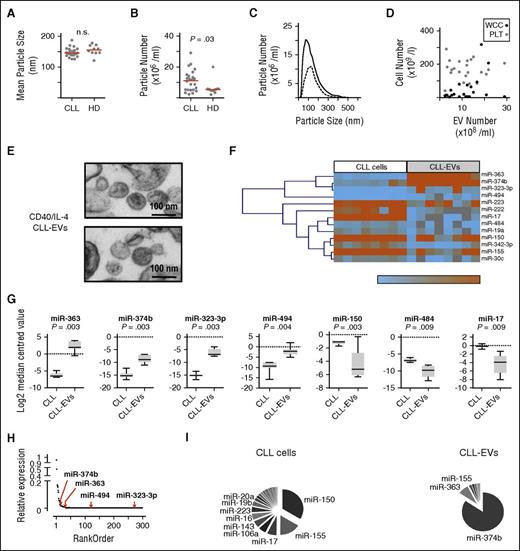
EVs from patient plasma (n = 23) were compared with age-matched healthy donors (n = 10) by dynamic light scattering (NanoSight; Malvern Instruments). Although the distribution of vesicle sizes was similar (Figure 1A), CLL patients showed significantly (P = .03) increased numbers of EVs compared with healthy donors (Figure 1B). The EV particle size distributions for CLL cases with higher or lower numbers of plasma vesicles were similar (Figure 1C) and EV concentration was not significantly correlated with absolute lymphocyte or platelet counts (Figure 1D). These findings are in agreement with previous studies14,15 and suggest that EV concentration in CLL does not simply reflect circulating tumor load.
EVs from CD40/IL-4–stimulated CLL cells are enriched for specific miRNAs. Dynamic light scattering (NanoSight) was used to compare (A) EV size between plasma of patients (CLL) (n = 23) and age-matched healthy donors (HD) (n = 10) and (B) EV particle number/concentration. EV numbers were significantly greater in CLL patients compared with HDs (Mann-Whitney test; P = .03). Horizontal bars represent median values. (C) Comparison of EV size distribution in 2 patients, 1 with high EV particle number (solid line) and the other with lower EV particle numbers (dashed line). (D) Lack of association between total white cell count (WCC; black circles) or platelet count (PLT; gray circles) and CLL-EV numbers. (E) Electron micrographs of CLL-EVs obtained after differential centrifugation of supernatant from CLL cells treated with anti-CD40 antibody (clone EA-5, 1 µg/mL), soluble CD40L (1 µg/mL), and IL-4 (20 ng/mL) for 36 hours. Top and bottom panels show representative CLL-EV particles obtained from 2 individual CLL patients. The size marker is 100 nm. (F) Unsupervised hierarchical clustering of miRNAs with significantly different expression between CLL-EVs and paired parental intracellular fractions from 8 CLL patients following CD40/IL-4 stimulation (Wilcoxon, Mann-Whitney test utilizing an FDR significance criterion limit of 0.05). (G) miRNAs that showed significant differences (Wilcoxon, Mann-Whitney test utilizing an FDR significance criterion limit of 0.01) in expression between CLL-EVs and paired parental intracellular fractions following CD40/IL-4 stimulation. Data are presented using box-and-whisker plots. miR-363, miR-374b, miR-323-3p, and miR-494 demonstrate increased expression in CLL-EVs whereas miR-150, miR-484, and miR-17 show reduced expression compared with parental CLL intracellular miRNA content. (H) Relative expression analysis plot. Cellular miRNA were ranked from the most highly expressed (relative expression arbitrarily set at 1) to the least expressed. The positions of the 4 miRNA whose expression level increased in CLL-EVs compared with parental CLL intracellular miRNA content is shown by the red arrows. The analysis shows that miRNAs that are highly expressed in CLL-EVs are a specific population and do not simply reflect the most highly expressed parental cellular miRNA content. (I) Pie charts showing relative distribution of miRNAs in parental CLL cells and paired CLL-EVs following CD40/IL-4 stimulation. The most highly expressed miRNAs in each compartment are labeled. n.s., not significant.
EVs from CD40/IL-4–stimulated CLL cells are enriched for specific miRNAs. Dynamic light scattering (NanoSight) was used to compare (A) EV size between plasma of patients (CLL) (n = 23) and age-matched healthy donors (HD) (n = 10) and (B) EV particle number/concentration. EV numbers were significantly greater in CLL patients compared with HDs (Mann-Whitney test; P = .03). Horizontal bars represent median values. (C) Comparison of EV size distribution in 2 patients, 1 with high EV particle number (solid line) and the other with lower EV particle numbers (dashed line). (D) Lack of association between total white cell count (WCC; black circles) or platelet count (PLT; gray circles) and CLL-EV numbers. (E) Electron micrographs of CLL-EVs obtained after differential centrifugation of supernatant from CLL cells treated with anti-CD40 antibody (clone EA-5, 1 µg/mL), soluble CD40L (1 µg/mL), and IL-4 (20 ng/mL) for 36 hours. Top and bottom panels show representative CLL-EV particles obtained from 2 individual CLL patients. The size marker is 100 nm. (F) Unsupervised hierarchical clustering of miRNAs with significantly different expression between CLL-EVs and paired parental intracellular fractions from 8 CLL patients following CD40/IL-4 stimulation (Wilcoxon, Mann-Whitney test utilizing an FDR significance criterion limit of 0.05). (G) miRNAs that showed significant differences (Wilcoxon, Mann-Whitney test utilizing an FDR significance criterion limit of 0.01) in expression between CLL-EVs and paired parental intracellular fractions following CD40/IL-4 stimulation. Data are presented using box-and-whisker plots. miR-363, miR-374b, miR-323-3p, and miR-494 demonstrate increased expression in CLL-EVs whereas miR-150, miR-484, and miR-17 show reduced expression compared with parental CLL intracellular miRNA content. (H) Relative expression analysis plot. Cellular miRNA were ranked from the most highly expressed (relative expression arbitrarily set at 1) to the least expressed. The positions of the 4 miRNA whose expression level increased in CLL-EVs compared with parental CLL intracellular miRNA content is shown by the red arrows. The analysis shows that miRNAs that are highly expressed in CLL-EVs are a specific population and do not simply reflect the most highly expressed parental cellular miRNA content. (I) Pie charts showing relative distribution of miRNAs in parental CLL cells and paired CLL-EVs following CD40/IL-4 stimulation. The most highly expressed miRNAs in each compartment are labeled. n.s., not significant.
To examine the contribution of immune-derived CD40 and IL-4 stimulation, we characterized EVs released from CLL cells following CD40 and IL-4 (CD40/IL-4 CLL-EVs).18,19 CD40/IL-4 stimulation provide signals for leukemia survival,19 in part through induction of antiapoptotic proteins and proliferation.23 To avoid confounding effects due to EVs produced by a feeder cell layer, we used soluble stimulators: anti-CD40 antibody, soluble CD40L, and IL-418 (supplemental Figure 1A-C). Unstimulated CLL-EVs (mean 134 nm, mode 110 nm) and CD40/IL-4 CLL-EVs (mean 150 nm, mode 100 nm) showed similar particle concentration and size distribution (supplemental Figure 1D-E). Although the CLL leukemia cell line MEC1 produced ∼15-fold more EVs (supplemental Figure 1F) compared with primary CLL cells, their particle size (mean 120 nm, mode 91 nm) was comparable to EVs from primary tumor cells.5 The tetraspanin protein CD9 was highly expressed on parental CLL cells and was detectable on CD40/IL-4 CLL-EVs, whereas CD63 was induced on both parental tumor cells and CLL-EVs following CD40/IL-4 stimulation (supplemental Figure 1G-H).
Next, we used low-density microarrays (Figure 1E-F) to compare miRNA profiles between EVs derived from CD40/IL-4–stimulated CLL cells and those of unstimulated parental cells (CLL). We found EV miRNA content to be less complex than that of cells5,24 (19 miRNAs in EVs compared with 136 in CLL cells). A signature of 13 (false discovery rate [FDR] <0.05) or 7 (FDR <0.01) miRNA that were differentially expressed between EV and cells was obtained: miR-363, miR-374b, miR-323-3p, and miR-494 were upregulated and miR-150, miR-484, and miR-17 were downregulated (Figure 1F-G). Ranking miRNA from the most highly to the least expressed showed that individual miRNA that were at relatively low levels in cells could be more highly expressed within EVs, indicating selective inclusion of miRNAs into EVs from the cellular pool (Figure 1H-I; supplemental Table 2). We validated the microarray findings by comparative reverse transcription PCR analyses of paired EVs and parental CLL cells with and without CD40/IL-4 stimulation (supplemental Figure 2). Enrichment of miR-363, miR-374b, miR-323-3p, and miR-494 in EVs compared with parental CLL cells was confirmed (supplemental Figure 2A). Although the amounts of these miRNA did not differ between unstimulated and stimulated parental CLL cells (supplemental Figure 2B), we detected significant enrichment of miR-363, miR-374b, miR-323-3p, and miR-494 in EVs derived from CD40/IL-4–stimulated CLL cells (CD40/IL-4 EVs) compared with EVs from unstimulated cells (CLL-EVs) (supplemental Figure 2C). Although some of the differentially expressed EV miRNAs (miR-155, miR-150, miR-191, and miR-223) identified from our miRNA profiling have been associated with CLL plasma-derived exosomes and BCR activation,14 or from cultured CLL cell-derived exosomes,16 our data show that CD40/IL-4 stimulation contributes to enrichment of specific miRNA within CLL-EVs
CLL-EVs carrying miR-363 can be functionally transferred to target CD4+ T cells
miR-363 showed ∼270-fold increased expression in EVs compared with parental CD40/IL-4–stimulated CLL cells (supplemental Table 2) and we identified the T-cell immunomodulatory protein CD6925 as having the highest context score of all miR-363 target genes (targetscan.org)26,27 (supplemental Table 3). Three potential conserved miR-363–binding sites are present in the CD69 3′ untranslated region (UTR) (Figure 2A-C) and luciferase reporter assays were carried out to confirm their functional role. Transfection of a miR-363 mimic significantly repressed CD69 luciferase activity, whereas mutation of all 3 miR-363–binding sites was required to maximally abolish repression of luciferase activity by miR-363 (Figure 2D; supplemental Table 4).
Transfer of CLL-EVs containing miR-363 to CD4+ T cells downregulates the expression of the immunomodulatory receptor CD69. (A) Potential binding sites for miR-363 in the 3′ UTR of human CD69 identified by TargetScan v7.0. (B) Diagram showing the position of the 3 miR-363–binding sites (labeled 1, 2, and 3) (TargetScan v7.0) in the CD69 3′ UTR. The 3′ UTR has a total length of 995 bp and site 1 is at 758 bp, site 2 at 870 bp, and site 3 at 907 bp. (C) Plots demonstrating conservation of miR-363–binding sites (horizontal black lines). Sequences from 6 species (human, mouse, dog, cat, cow, and rabbit) were compared (weblogo.berkely.edu). The overall height of the stack indicates the sequence conservation at that position, whereas the height of symbols within the stack indicates the relative frequency of each amino or nucleic acid at that position. (D) Luciferase activity, in HEK293T cells, following cotransfection of reporter constructs with either miR-363 mimic or scrambled control miRNA expressed as percentage of luciferase activity following transfection of the reporter construct alone. Five reporter constructs were tested: wild-type (WT), construct-bearing mutations of site 1 (labeled 1), construct-bearing mutations of site 2 (labeled 2), construct-bearing mutations of site 3 (labeled 3), and construct-bearing mutations of sites 1, 2, and 3 (labeled 1, 2, 3). The miR-363 mimic caused repression of luciferase activity compared with the scrambled control unless all 3 miR-363 sites were mutated (n = 4). Mean ± SEM. The 2-tailed Student t test was used (***P < .001, **P < .01, *P < .05). (E) Flow cytometry histograms showing repression of CD69 expression in the human Jurkat T cells following transfection with miR-363 mimic. T cells were either transfected with scrambled control miRNA (solid blue line) or miR-363 mimic (solid red line) and stimulated with PMA/ionomycin. FACS histograms for control unstimulated cells are shown as red and blue dotted lines. Bar chart shows that miR-363 mimic caused a significant (Student t test, *P < .05) reduction in MFI (n = 4). (F) Incubation of primary CD4+ T cells with autologous CLL-EVs following CD40/IL-4 stimulation increased intracellular levels (relative change) of T-cell miR-363. Mean ± SEM from 3 separate experiments are shown (Student t test, **P < .01). (G) Flow cytometry analysis revealing that incubation of primary CD4+ T cells with autologous CLL-EVs following CD40/IL-4 stimulation (red line) led to significant suppression of CD69 expression following crosslinking of T cells with anti-CD3/CD28 compared with the treatment of T cells with control CLL-EVs from unstimulated CLL cells (blue line). Dotted lines are the respective isotype control experiments. The bar chart shows changes to MFI expression of CD69 following treatment with CLL-EVs (Student t test, *P < .05) (n = 4). FACS, fluorescence-activated cell sorter; MFI, mean fluorescence intensity; n.s., not significant; PE, phycoerythrin; Scr, scrambled; SEM, standard error of the mean.
Transfer of CLL-EVs containing miR-363 to CD4+ T cells downregulates the expression of the immunomodulatory receptor CD69. (A) Potential binding sites for miR-363 in the 3′ UTR of human CD69 identified by TargetScan v7.0. (B) Diagram showing the position of the 3 miR-363–binding sites (labeled 1, 2, and 3) (TargetScan v7.0) in the CD69 3′ UTR. The 3′ UTR has a total length of 995 bp and site 1 is at 758 bp, site 2 at 870 bp, and site 3 at 907 bp. (C) Plots demonstrating conservation of miR-363–binding sites (horizontal black lines). Sequences from 6 species (human, mouse, dog, cat, cow, and rabbit) were compared (weblogo.berkely.edu). The overall height of the stack indicates the sequence conservation at that position, whereas the height of symbols within the stack indicates the relative frequency of each amino or nucleic acid at that position. (D) Luciferase activity, in HEK293T cells, following cotransfection of reporter constructs with either miR-363 mimic or scrambled control miRNA expressed as percentage of luciferase activity following transfection of the reporter construct alone. Five reporter constructs were tested: wild-type (WT), construct-bearing mutations of site 1 (labeled 1), construct-bearing mutations of site 2 (labeled 2), construct-bearing mutations of site 3 (labeled 3), and construct-bearing mutations of sites 1, 2, and 3 (labeled 1, 2, 3). The miR-363 mimic caused repression of luciferase activity compared with the scrambled control unless all 3 miR-363 sites were mutated (n = 4). Mean ± SEM. The 2-tailed Student t test was used (***P < .001, **P < .01, *P < .05). (E) Flow cytometry histograms showing repression of CD69 expression in the human Jurkat T cells following transfection with miR-363 mimic. T cells were either transfected with scrambled control miRNA (solid blue line) or miR-363 mimic (solid red line) and stimulated with PMA/ionomycin. FACS histograms for control unstimulated cells are shown as red and blue dotted lines. Bar chart shows that miR-363 mimic caused a significant (Student t test, *P < .05) reduction in MFI (n = 4). (F) Incubation of primary CD4+ T cells with autologous CLL-EVs following CD40/IL-4 stimulation increased intracellular levels (relative change) of T-cell miR-363. Mean ± SEM from 3 separate experiments are shown (Student t test, **P < .01). (G) Flow cytometry analysis revealing that incubation of primary CD4+ T cells with autologous CLL-EVs following CD40/IL-4 stimulation (red line) led to significant suppression of CD69 expression following crosslinking of T cells with anti-CD3/CD28 compared with the treatment of T cells with control CLL-EVs from unstimulated CLL cells (blue line). Dotted lines are the respective isotype control experiments. The bar chart shows changes to MFI expression of CD69 following treatment with CLL-EVs (Student t test, *P < .05) (n = 4). FACS, fluorescence-activated cell sorter; MFI, mean fluorescence intensity; n.s., not significant; PE, phycoerythrin; Scr, scrambled; SEM, standard error of the mean.
To show functional importance of miR-363, we transfected Jurkat T cells with either a miR-363 mimic or a scrambled control miRNA prior to phorbol myristate acetate (PMA)/ionomycin stimulation to induce CD69 expression. Reverse transcription PCR for miR-363 demonstrated increased levels within Jurkat T cells following transfection (supplemental Figure 3A) and this associated with significantly repressed PMA/ionomycin-induced expression of CD69 (P < .05) (Figure 2E). CD69 expression was inducible on primary CD4+ T cells following crosslinking with anti-CD3/CD28. Next, primary CD4+ T cells were cultured with CLL-EVs following CD40/IL-4 stimulation (CD40/IL-4 CLL-EVs) compared with control EVs from unstimulated CLL cells (CLL-EVs) in order to determine their effect on CD69 expression. CD40/IL-4 CLL-EVs increased expression levels of miR-363 in target T cells (P < .01) (Figure 2F) and led to significant suppression of CD69 expression following crosslinking (P < .05) (Figure 2G). To confirm uptake of CLL-EVs by primary T cells, transfer coculture experiments were carried out and demonstrated maximal uptake of fluorescently (PKH67) labeled CD40/IL-4 CLL-EVs into autologous CD4+ T cells by 24 hours (Figure 3). Taken together, we show that EVs released from CD40/IL-4–stimulated CLL cells that carry miR-363 can be functionally transferred to target CD4+ T cells, resulting in direct modulation of the mRNA target CD69.
EVs from CD40/IL-4–stimulated CLL cells rapidly enter autologous CD4+ T cells. CD4+ T cells from CLL patients were incubated with fluorescently PKH67-labeled CD40/IL-4 CLL-EVs for the indicated times before washing and fixation (0 time point, 24 and 48 hours). EV transfer studies were followed by confocal imaging. (A) Quantification of EV internalization using ImageJ software. MFI of PKH67-EVs ± SEM (n = 3). **P < .01. (B) The confocal images show maximal uptake of PKH67-labeled CLL-EVs into autologous CD4+ T cells by 24 hours (CD4+ T-cell F-actin, rhodamine phalloidin [red], PKH67-labeled CD40/IL-4 CLL-EVs [green], CD4+ T-cell nuclei [blue], and a merged image [bottom right of image panels]). Scale bar, 20 μm. (C) Confocal images showing serial sections of confocal micrographs (CD4+ T-cell F-actin, rhodamine phalloidin [red] and PKH67-labeled CD40/IL-4 CLL-EVs [green]) showing internalization of CLL-EVs within CD4+ T cells by 24 hours. Scale bar, 5 μm.
EVs from CD40/IL-4–stimulated CLL cells rapidly enter autologous CD4+ T cells. CD4+ T cells from CLL patients were incubated with fluorescently PKH67-labeled CD40/IL-4 CLL-EVs for the indicated times before washing and fixation (0 time point, 24 and 48 hours). EV transfer studies were followed by confocal imaging. (A) Quantification of EV internalization using ImageJ software. MFI of PKH67-EVs ± SEM (n = 3). **P < .01. (B) The confocal images show maximal uptake of PKH67-labeled CLL-EVs into autologous CD4+ T cells by 24 hours (CD4+ T-cell F-actin, rhodamine phalloidin [red], PKH67-labeled CD40/IL-4 CLL-EVs [green], CD4+ T-cell nuclei [blue], and a merged image [bottom right of image panels]). Scale bar, 20 μm. (C) Confocal images showing serial sections of confocal micrographs (CD4+ T-cell F-actin, rhodamine phalloidin [red] and PKH67-labeled CD40/IL-4 CLL-EVs [green]) showing internalization of CLL-EVs within CD4+ T cells by 24 hours. Scale bar, 5 μm.
Autologous CD4+ T cells exposed to EVs from CD40/IL-4–stimulated CLL cells exhibit enhanced immunological synapse signaling and migration
Given the link between CLL cells and CD4+ T cells within TME proliferation centers,7,19 we hypothesized that EVs from CD40/IL-4–stimulated CLL cells might influence T-cell function. Treatment of autologous CD4+ T cells with CD40/IL-4 CLL-EVs significantly enhanced F-actin immunological synapse formation with CLL tumor cells compared with control CLL-EVs (P < .01) (Figure 4A). Transwell culture assays confirmed that the transfer of secreted particles from CD40/IL-4–stimulated CLL cells modulated T-cell immune synapse function compared with unstimulated CLL cells (data not shown). In addition, we observed that the percentage efficiency of CD4+ T cells to form conjugates was increased (P < .05) following exposure to CD40/IL-4 CLL-EVs compared with control CLL-EVs (Figure 4B). Next, we examined the recruitment/polarization of proximal T-cell receptor–mediated tyrosine kinases to CD4+ T-cell immune synapses following treatment with CLL-EVs. Our results showed significantly increased recruitment of Lck (P < .01) (Figure 4C) and tyrosine-phosphorylated proteins (data not shown) to the T-cell:CLL synapse contact sites following exposure to CD40/IL-4 CLL-EVs compared with CLL-EVs. Notably, treatment of CD4+ T cells with CD40/IL-4 CLL-EVs augmented immune synapse activity while downregulating CD69 expression levels on T cells compared with CLL-EVs (P < .01) (Figure 4D). To validate the ability of CD40/IL-4 CLL-EVs to modulate T-cell synapse signaling, we knocked down Rab27A/B in parental CLL cells using locked nucleic acid (LNA)-based antisense oligonucleotides (supplemental Figure 4A) prior to CD40/IL-4 stimulation and isolation of released EVs Rab27A/B participates in the traffic of EVs, including exosomes to the cell periphery as part of the secretory pathway.11 We found that Rab27A/B knockdown in CLL cells impaired the release of EVs (supplemental Figure 4B), which prevented modulation of synapse signaling (supplemental Figure 4C). CD40/IL-4 CLL-EV–mediated increased synapse formation with enhanced early CD4+ T-cell signaling would be expected to augment T-cell proliferation. To investigate this, we measured nuclear Ki-67 expression, a marker for cellular proliferation, in autologous patient CD4+ T cells exposed to both CLL-EVs and anti-CD3/-CD28 crosslinking. As shown in Figure 4E-G, coculture with CD40/IL-4 CLL-EVs increased Ki-67 expression in CD4+ T cells compared with coculture with control CLL-EVs. Carboxyfluorescein diacetate succinimidyl ester (CFSE)-labeled CD4+ T cells confirmed that exposure to CD40/IL-4 CLL-EVs enhanced T-cell proliferation (data not shown).
CD4+ T cells exposed to autologous EVs from CD40/IL-4–stimulated CLL cells exhibit enhanced immunological synapse signaling and proliferation. (A) Confocal images of T-cell F-actin immune synapse formation (F-actin rhodamine phalloidin, red) interactions with autologous tumor CLL cells (CMAC-labeled, blue) from a representative patient. CD4+ T cells (negatively selected) were treated with purified CLL-EVs for 48 hours (CLL-EVs following CD40/IL-4 stimulation [CD40/IL-4 CLL-EVs]) compared with control EVs from unstimulated CLL cells (CLL-EVs). Treated T cells were then conjugated with autologous CLL B cells (pulsed with superantigen, sAg cocktail and labeled with CellTracker blue dye, CMAC). T-cell conjugates were analyzed by immunofluorescence and confocal microscopy. Confocal images (n = 10 per patient treatment sample) were acquired using an A1R confocal microscope (Nikon) and analyzed with NIS-Elements software (Nikon). Original magnification, ×60. Scale bar, 5 μm. Quantitative image analysis measured the total area (μm2) of F-actin polymerization (F-actin rhodamine phalloidin, red) at CD4+ T-cell contact sites and synapses with CLL cells within the experimental T-cell population. The dot plot shows the mean synapse/interaction area (μm2) ± SEM from a representative patient (from 6 independent CLL samples) (Mann-Whitney t test, **P < .01). (B) Intercellular CD4+ T cell: tumor CLL conjugates formed between CD40/IL-4 CLL-EVs or CLL-EVs treated CD4+ T cells and sAg-pulsed autologous CLL tumor cells were scored using a confocal microscope (scale bar, 5 μm) and image analysis software. Bar chart shows the mean percentage CD4+ T-cell:tumor CLL conjugates ± SEM from 6 independent CLL samples with 100 random T cells analyzed per experiment (Mann-Whitney t test, *P < .05). (C) CD4+ T-cell:tumor CLL conjugates formed during 20 minutes were fixed, permeabilized, and stained with anti-Lck (green). Tumor cells were labeled with CellTracker blue dye, CMAC. T cells are denoted by white dashed lines. Quantitative image analysis (relative recruitment index [RRI]) of Lck accumulation at the immunological synapse is shown, representative evaluation of 20 conjugates from 3 independent experiments, with the mean value shown as a blue bar ± SEM. Scale bar, 5 μm. (D) MFI was calculated for CD4+ T-cell CD69 expression levels (Alexa 488, green fluorescent channel) following treatment with CD40/IL-4 CLL-EVs or CLL-EVs T cells are denoted by white dashed lines. Tumor cells were labeled with CellTracker blue dye, CMAC. Dot plot chart shows the CD69 MFI ± SEM for a representative patient from 6 CLL samples (Mann-Whitney t test, **P < .01). Original magnification, ×60. Scale bar, 5 μm. (E) Nuclear Ki-67 expression, a marker for cellular proliferation, was measured in autologous patient CD4+ T cells exposed to CLL-EVs (CD40/IL-4 CLL-EVs or CLL-EVs) and with and without anti-CD3/-CD28 crosslinking stimulation for 72 hours (-CD3/-CD28 and unstim., respectively). Bar chart shows mean Ki-67 ± SEM for 3 independent CLL patient experiments (1-way ANOVA, **P < .01). (F) The confocal images show increased nuclear (DAPI, blue) Ki-67 expression (Alexa 647 denoted using green with analysis software) in CD4+ T cells (F-actin, red) treated with CD40/IL-4 CLL-EVs compared with coculture with control CLL-EVs. Scale bar, 5 μm. (G) Z-stack confocal analysis (3-dimensional central T-cell region analysis) reveals increased nuclear (DAPI, blue) Ki-67 expression (Alexa 647 denoted using green with analysis software) in CD4+ T cells (F-actin, red) treated with CD40/IL-4 CLL-EVs compared with control CLL-EVs. Scale bar, 5 μm. ANOVA, analysis of variance; DAPI, 4′,6-diamidino-2-phenylindole.
CD4+ T cells exposed to autologous EVs from CD40/IL-4–stimulated CLL cells exhibit enhanced immunological synapse signaling and proliferation. (A) Confocal images of T-cell F-actin immune synapse formation (F-actin rhodamine phalloidin, red) interactions with autologous tumor CLL cells (CMAC-labeled, blue) from a representative patient. CD4+ T cells (negatively selected) were treated with purified CLL-EVs for 48 hours (CLL-EVs following CD40/IL-4 stimulation [CD40/IL-4 CLL-EVs]) compared with control EVs from unstimulated CLL cells (CLL-EVs). Treated T cells were then conjugated with autologous CLL B cells (pulsed with superantigen, sAg cocktail and labeled with CellTracker blue dye, CMAC). T-cell conjugates were analyzed by immunofluorescence and confocal microscopy. Confocal images (n = 10 per patient treatment sample) were acquired using an A1R confocal microscope (Nikon) and analyzed with NIS-Elements software (Nikon). Original magnification, ×60. Scale bar, 5 μm. Quantitative image analysis measured the total area (μm2) of F-actin polymerization (F-actin rhodamine phalloidin, red) at CD4+ T-cell contact sites and synapses with CLL cells within the experimental T-cell population. The dot plot shows the mean synapse/interaction area (μm2) ± SEM from a representative patient (from 6 independent CLL samples) (Mann-Whitney t test, **P < .01). (B) Intercellular CD4+ T cell: tumor CLL conjugates formed between CD40/IL-4 CLL-EVs or CLL-EVs treated CD4+ T cells and sAg-pulsed autologous CLL tumor cells were scored using a confocal microscope (scale bar, 5 μm) and image analysis software. Bar chart shows the mean percentage CD4+ T-cell:tumor CLL conjugates ± SEM from 6 independent CLL samples with 100 random T cells analyzed per experiment (Mann-Whitney t test, *P < .05). (C) CD4+ T-cell:tumor CLL conjugates formed during 20 minutes were fixed, permeabilized, and stained with anti-Lck (green). Tumor cells were labeled with CellTracker blue dye, CMAC. T cells are denoted by white dashed lines. Quantitative image analysis (relative recruitment index [RRI]) of Lck accumulation at the immunological synapse is shown, representative evaluation of 20 conjugates from 3 independent experiments, with the mean value shown as a blue bar ± SEM. Scale bar, 5 μm. (D) MFI was calculated for CD4+ T-cell CD69 expression levels (Alexa 488, green fluorescent channel) following treatment with CD40/IL-4 CLL-EVs or CLL-EVs T cells are denoted by white dashed lines. Tumor cells were labeled with CellTracker blue dye, CMAC. Dot plot chart shows the CD69 MFI ± SEM for a representative patient from 6 CLL samples (Mann-Whitney t test, **P < .01). Original magnification, ×60. Scale bar, 5 μm. (E) Nuclear Ki-67 expression, a marker for cellular proliferation, was measured in autologous patient CD4+ T cells exposed to CLL-EVs (CD40/IL-4 CLL-EVs or CLL-EVs) and with and without anti-CD3/-CD28 crosslinking stimulation for 72 hours (-CD3/-CD28 and unstim., respectively). Bar chart shows mean Ki-67 ± SEM for 3 independent CLL patient experiments (1-way ANOVA, **P < .01). (F) The confocal images show increased nuclear (DAPI, blue) Ki-67 expression (Alexa 647 denoted using green with analysis software) in CD4+ T cells (F-actin, red) treated with CD40/IL-4 CLL-EVs compared with coculture with control CLL-EVs. Scale bar, 5 μm. (G) Z-stack confocal analysis (3-dimensional central T-cell region analysis) reveals increased nuclear (DAPI, blue) Ki-67 expression (Alexa 647 denoted using green with analysis software) in CD4+ T cells (F-actin, red) treated with CD40/IL-4 CLL-EVs compared with control CLL-EVs. Scale bar, 5 μm. ANOVA, analysis of variance; DAPI, 4′,6-diamidino-2-phenylindole.
Rapid motility is essential for the trafficking of T cells into lymphoid tissue.28 Video microscopy with tracking analysis was then used to assess the migration of CLL-EV–treated autologous CD4+ T cells on ICAM-1–coated plates using the chemokine CCL19, which attracts T cells within the lymph node.22,28 Our data show that treatment of CD4+ T cells with CD40/IL-4 CLL-EVs significantly increased migration rates (both speed and distance) compared with control CLL-EV–treated T cells (paired t test, P < .01) (Figure 5 and supplemental Videos 1-2, respectively).
CD4+ T cells exposed to autologous EVs from CD40/IL-4–stimulated CLL cells exhibit enhanced CCL19-induced migration. Primary patient CD4+ T cells were treated with autologous CLL-EVs for 48 hours (CLL-EVs following CD40/IL-4 stimulation [CD40/IL-4 CLL-EVs], compared with control EVs from unstimulated CLL cells [CLL-EVs]). Treated T cells were then allowed to adhere to immobilized ICAM-1 for 10 minutes following exposure to the chemokine CCL19. Video microscopy (Biostation IM; Nikon) was used to observe the migration of CD4+ T cells for 20 minutes before the migration of individual cells was tracked (NIS-Elements analysis software; Nikon). (A) Representative phase-contrast images (original magnification, ×20) and motility tracks (colored) of CD4+ T cells from CLL patients treated with experimental CLL-EVs as denoted (top microscopy images). Representative migratory tracks of individual T cells from CLL patients treated with CLL-EVs are shown in the bottom data plots (n = 20 cells per experiment). Dot plot charts show the (B) speed of migration (micromolar per minute) and (C) distance of migration (μm) for a representative patient from 6 independent CLL samples (Mann-Whitney t test, **P < .01).
CD4+ T cells exposed to autologous EVs from CD40/IL-4–stimulated CLL cells exhibit enhanced CCL19-induced migration. Primary patient CD4+ T cells were treated with autologous CLL-EVs for 48 hours (CLL-EVs following CD40/IL-4 stimulation [CD40/IL-4 CLL-EVs], compared with control EVs from unstimulated CLL cells [CLL-EVs]). Treated T cells were then allowed to adhere to immobilized ICAM-1 for 10 minutes following exposure to the chemokine CCL19. Video microscopy (Biostation IM; Nikon) was used to observe the migration of CD4+ T cells for 20 minutes before the migration of individual cells was tracked (NIS-Elements analysis software; Nikon). (A) Representative phase-contrast images (original magnification, ×20) and motility tracks (colored) of CD4+ T cells from CLL patients treated with experimental CLL-EVs as denoted (top microscopy images). Representative migratory tracks of individual T cells from CLL patients treated with CLL-EVs are shown in the bottom data plots (n = 20 cells per experiment). Dot plot charts show the (B) speed of migration (micromolar per minute) and (C) distance of migration (μm) for a representative patient from 6 independent CLL samples (Mann-Whitney t test, **P < .01).
Knockdown of miR-363 in CLL cells prevents the ability of released CLL-EVs to confer altered functional properties to CD4+ T cells
To determine whether CLL-EV miR-363 is a significant regulator of CD4+ T-cell biology, we knocked down expression of this miR in CLL cells using LNA-based antisense oligonucleotides, prior to CD40/IL-4 stimulation and isolation of released EVs (supplemental Figure 3B). miR-363 knockdown prevented the ability of CD40/IL-4 CLL-EVs to confer increased F-actin immune synapse polarization at CD4+ T-cell contact sites with autologous CLL tumor cells (Figure 6A). Moreover, miR-363 knockdown prevented enhanced tyrosine kinase recruitment to the synapse site (Figure 6B) compared with CD40/IL-4 CLL-EVs derived from CLL transfected with scrambled control antisense oligonucleotides. Critically, CD69 expression levels remained unaffected in CD4+ T cells following exposure to CD40/IL-4 CLL-EVs harvested from miR-363 knocked down CLL cells (Figure 6C), providing further evidence that the immunomodulatory receptor CD69 is a target of CD40/IL-4 CLL-EV miR-363.
Knockdown of miR-363 in parental CLL cells prevents the ability of released CLL-EVs to confer altered functional properties to autologous CD4+ T cells. miR-363 was knocked down in CLL cells using LNA-based antisense oligonucleotides, prior to CD40/IL-4 stimulation and isolation of released EVs (anti-miR-363 compared with scrambled control [Con.] antisense miR oligonucleotides). CD4+ T cells (negatively selected) were treated with purified CLL-EVs from the transfected autologous parental CLL cells. Treated T cells were then conjugated with autologous CLL B cells (pulsed with superantigen [sAg] cocktail and labeled with CellTracker blue dye, CMAC). T-cell conjugates were analyzed by immunofluorescence and confocal microscopy as described in Figure 4. Confocal images (n = 10 per patient treatment sample) were acquired using an A1R confocal microscope (Nikon) and analyzed with NIS-Elements software (Nikon). Original magnification, ×60. Scale bar, 5 μm. Dot plot charts show (A) the mean F-actin polymerization (rhodamine phalloidin, red) synapse/interaction area (μm2) ± SEM from a representative patient (from 3 independent CLL samples), (B) the mean RRI of Lck accumulation (green) at the immunological synapse, representative analysis (±SEM) from 3 independent patient experiments and (C) the CD69 MFI ± SEM for a representative patient from 3 independent CLL patient samples. CD69 expression is denoted in white. Treatment of autologous CD4+ T cells with CLL-EVs derived from miR-363 knocked down cells (anti-miR-363) blocked modulation of T-cell migration rates when compared with EVs derived from control transfected tumor cells (Scrambled Con. miR). Dot plot charts show the (D) speed of migration (micromolar per minute) and (E) distance of migration (μm) for a representative patient from 3 independent CLL samples (Mann-Whitney t test, *P < .05, **P < .01).
Knockdown of miR-363 in parental CLL cells prevents the ability of released CLL-EVs to confer altered functional properties to autologous CD4+ T cells. miR-363 was knocked down in CLL cells using LNA-based antisense oligonucleotides, prior to CD40/IL-4 stimulation and isolation of released EVs (anti-miR-363 compared with scrambled control [Con.] antisense miR oligonucleotides). CD4+ T cells (negatively selected) were treated with purified CLL-EVs from the transfected autologous parental CLL cells. Treated T cells were then conjugated with autologous CLL B cells (pulsed with superantigen [sAg] cocktail and labeled with CellTracker blue dye, CMAC). T-cell conjugates were analyzed by immunofluorescence and confocal microscopy as described in Figure 4. Confocal images (n = 10 per patient treatment sample) were acquired using an A1R confocal microscope (Nikon) and analyzed with NIS-Elements software (Nikon). Original magnification, ×60. Scale bar, 5 μm. Dot plot charts show (A) the mean F-actin polymerization (rhodamine phalloidin, red) synapse/interaction area (μm2) ± SEM from a representative patient (from 3 independent CLL samples), (B) the mean RRI of Lck accumulation (green) at the immunological synapse, representative analysis (±SEM) from 3 independent patient experiments and (C) the CD69 MFI ± SEM for a representative patient from 3 independent CLL patient samples. CD69 expression is denoted in white. Treatment of autologous CD4+ T cells with CLL-EVs derived from miR-363 knocked down cells (anti-miR-363) blocked modulation of T-cell migration rates when compared with EVs derived from control transfected tumor cells (Scrambled Con. miR). Dot plot charts show the (D) speed of migration (micromolar per minute) and (E) distance of migration (μm) for a representative patient from 3 independent CLL samples (Mann-Whitney t test, *P < .05, **P < .01).
We further show that treatment of autologous CD4+ T cells with CD40/IL-4 CLL-EVs derived from miR-363 knocked down cells blocked modulation of T-cell migration rates when compared with CD40/IL-4 CLL-EVs derived from control transfected tumor cells (Figure 6D-E and supplemental Videos 3-4, respectively). These data together identify the capacity of CD40/IL-4 CLL-EV miR-363 to play a critical role in regulating T-cell motility and immune synapse signaling function.
Discussion
Complex bidirectional crosstalk between CLL tumor cells and nonmalignant cells within the TME plays a critical role in activating tumor survival, proliferation, and fostering immune privilege.3,29 T cells from CLL patients exhibit impaired F-actin immunological synapse formation that contributes to reduced activation and effector functions.21 CLL tumor cells can suppress proximal activated antitumor T-cell activity by delivering inhibitory signals via co-option of the immune checkpoint network including the PD-L1:PD-1 pathway to dampen formation of the lytic immune synapse.6 Immune checkpoint blockade immunotherapy (including anti-PD-1 and anti-PD-L1 antibody drugs) has the potential to activate T cells and unleash antitumor immune responses, serving as an illustrative example of therapeutically targeting/reeducating the TME. However, there is evidence that subverted CD4+ T-cell subsets within the lymphoid TME may exhibit tumor-promoting activity that complicates our understanding of cancer immunology and raises the issue that immunotherapy might have negative as well as positive activity.4 Investigating multifaceted bidirectional tumor–T-cell crosstalk will be essential to understand both disease pathogenesis and the effects of novel immunotherapies within the TME.
CLL cells proliferate in distinct microanatomical tissue sites termed proliferation centers (pseudofollicles) that allow intimate interactions with TME components including activated CD4+ T cells.1 Immunohistochemistry analysis demonstrated that CLL cells in proliferation centers are in close proximity to CD4+ T cells.30 More recently, imaging studies have revealed that proliferating CLL cells were found to colocalize and form intercellular conjugates with activated CD4+ T cells in lymph node tissue.7 Activated CD4+ T cells including follicular T helper cells cells can provide CD40L costimulation and are a source of prosurvival cytokines (IL-4 and IL-21) that activate extracellular signal-regulated kinase, STAT, and NF-κB signaling and CLL proliferation.31-34 Moreover, a key role for CD4+ T cells in promoting CLL cell survival and expansion has been identified using a humanized model of disease. Activated CD4+ T cells were found to localize in pseudofollicular structures (proliferation center-like) and surround CLL tumor cells within murine lymphoid TME tissue.35
Among the mechanisms by which cells communicate within the TME, emerging evidence suggests that EVs released from tumor cells have the ability to modulate the microenvironment to promote immune evasion, angiogenesis, and stromal cell activation.13,36 It is believed that EVs have an important role in intercellular communication as they have shown the potential to transfer their contents including proteins, lipids, and RNAs (including mRNA and miRNA) between cells.13,37 Early studies focused on the ability of cancer cell-derived EVs that express tumor antigens to activate dendritic cells as a means to elicit antitumor immunity. However, there is now mounting evidence suggesting predominantly immunosuppressive functions for tumor EVs,13 including the induction of T-cell apoptosis,38 suppression of T-cell receptor signaling, and promoting the activity of T regulatory cells39 or tolerogenic dendritic cells.40
miRNAs (miRs) are important components of the molecular cargo of EVs and recent work has identified a disease-relevant EV microRNA profile in CLL.14,16 In particular, work has revealed the influence BCR signaling has on the release of CLL EVs and the inclusion of selective miR content including miR-150 and miR-155.14 The potential of CLL cell-derived EVs to modulate and reeducate the host microenvironment has been demonstrated with studies showing that CLL-EVs and their molecular cargo can be actively transferred to stromal cells inducing intracellular signaling and the reprogramming of these cells into proinflammatory cancer-associated fibroblast-like cells.5,15 However, the effects of CLL-EVs on T cells have not been previously described.
In the present study, we aimed to characterize EVs released from primary human CLL cells in response to the T-cell–derived TME signals CD40 and IL-4. These extracellular stimuli are known to produce prosurvival and proliferative signals in CLL.23,41 We report on the miR profiling of CLL-EVs derived from CLL cells stimulated with CD40 and IL-4 and reveal an enrichment of specific cellular miRNAs including miR-363 compared with parental cell miRNA content and control EVs from unstimulated CLL cells. Our data show that CD40/IL-4 stimulation contributes to the inclusion of selective miRNA within CLL-EVs distinct from BCR activation5,14 or cultured CLL cell-derived exosomes.16
We demonstrate that autologous patient CD4+ T cells internalize CLL-EVs containing miR-363 that targets the immunomodulatory receptor CD69. Our data support the functional transfer of this miR to target CD4+ T cells. Bioinfomatic analysis revealed that CD69 was a target gene for miR-363, one of the most highly enriched miRNAs in CLL-EVs released from CD40/IL-4–stimulated CLL cells (∼270-fold increased expression in EVs compared with parental CLL cells). CD69 is an early T-cell activation molecule expressed at sites of chronic inflammation.25 The precise immunomodulatory function of CD69 has not been elucidated owing to the absence of a known ligand and adequate in vivo models to study its physiological function. Although early data suggest that CD69 acts as a costimulatory receptor, recent studies have suggested a more complex nonredundant role for this receptor in the downregulation of T-cell responses,25,42 including inhibiting the migration of effector T cells.43,44
Our functional studies reveal that exposure of CD4+ T cells to EVs from CD40/IL-4–stimulated CLL cells enhances their migration, immunological synapse signaling, interactions with autologous tumor cells, and proliferation. Immune synapses are cellular signaling structures that orchestrate the complex communication between the T-cell and an antigen-presenting cell in a way that ensures detailed antigen recognition and effective T responses22 (and potentially bidirectional cooperative activation signaling to B cells).45 Notably, treatment of CD4+ T cells with CLL-EVs augmented immune synapse activity while downregulating CD69 expression levels on T cells. Intriguingly, knockdown of miR-363 in CLL cells using LNA antisense oligonucleotides prior to CD40/IL-4 stimulation prevented the ability of CLL-EVs to induce increased synapse signaling and confer altered functional properties to CD4+ T cells, suggesting an unsuspected and important role for this specific miRNA in regulating T-cell function. CD69 expression levels remained unaffected in these CD4+ T cells exposed to miR-363 knocked down CLL-EVs compared with control antisense oligonucleotides, providing evidence that T-cell CD69 is a target of transferred EV miR-363. These data support an important role for CD40/IL-4 CLL-EV–derived miR-363 in modulating CD4+ T-cell activity. In addition, these findings link the regulation of the immunomodulatory receptor CD69 to the miRNA cargo of CLL-EVs and suggest that CD69 may have a potential negative regulator function in CLL patient CD4+ T cells. Clearly, future studies will need to determine the precise roles of miR-363 and CD69 in generating the EV-induced T-cell functional effects we have observed. miR-363 levels in CLL patients’ plasma correlates with disease stage and some prognostic factors.46 Together with our data, this supports a rationale for further studies investigating the potential of miR-363 to act as a TME-derived biomarker for disease biology and response to therapy including immunotherapy. It is likely that miR-363 is not the only miRNA capable of repressing CD69 and indeed miR-374b, which we also found to be enriched in our CLL-EVs, has a potential binding site at bases 26 to 32 in the 3′ UTR of CD69.
Taken together, these data reveal a novel role for CLL-EVs in modifying CD4+ T-cell function that highlights unanticipated complexity of intercellular communication within the TME. The capacity of CLL-EVs and their specific molecular cargo (released following the T-cell–derived signals CD40 and IL-4) to enhance CD4+ T-cell migration and immune synapse interactions with autologous tumor cells may represent a novel immune-regulatory mechanism in the CLL TME.
The microarray data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE54434).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank David Dinsdale (Medical Research Council [MRC] Toxicology Unit, Leicester) for electron microscopy and Alison Goodall (University of Leicester) for use of the NanoSight machine. The authors also thank the Nikon Imaging Facility at King's College London for use of the Point Scanning Confocal A1R microscope, and all facility staff (Daniel Matthews, Isma Ali and Daniel Metcalf) who provided support.
This work was supported by a grant from the Kay Kendall Leukaemia Fund (KKL502; S.D.W.), and grants from the British Society of Haematology (BSH) and Bloodwise (14025; A.G.R.).
Authorship
Contribution: D.T.S., S.W., B.A., A.G.R., and S.D.W. designed experiments; D.T.S., B.A., S.W., L.L., A.A., A.R.A., and G.D.R. carried out experiments and analyzed data; and A.G.R. and S.D.W. supervised the experiments, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Simon D. Wagner, Room 104, Hodgkin Building, University of Leicester, Lancaster Rd, Leicester LE1 7HB, United Kingdom; e-mail: sw227@le.ac.uk; or Alan G. Ramsay, Department of Haemato-Oncology, Division of Cancer Studies, The Rayne Institute, King’s College London, 123 Coldharbour Ln, London, SE5 9NU, United Kingdom; e-mail: alan.ramsay@kcl.ac.uk.
References
Author notes
D.T.S., B.A., A.G.R., and S.D.W. contributed equally to this work.


![Figure 3. EVs from CD40/IL-4–stimulated CLL cells rapidly enter autologous CD4+ T cells. CD4+ T cells from CLL patients were incubated with fluorescently PKH67-labeled CD40/IL-4 CLL-EVs for the indicated times before washing and fixation (0 time point, 24 and 48 hours). EV transfer studies were followed by confocal imaging. (A) Quantification of EV internalization using ImageJ software. MFI of PKH67-EVs ± SEM (n = 3). **P < .01. (B) The confocal images show maximal uptake of PKH67-labeled CLL-EVs into autologous CD4+ T cells by 24 hours (CD4+ T-cell F-actin, rhodamine phalloidin [red], PKH67-labeled CD40/IL-4 CLL-EVs [green], CD4+ T-cell nuclei [blue], and a merged image [bottom right of image panels]). Scale bar, 20 μm. (C) Confocal images showing serial sections of confocal micrographs (CD4+ T-cell F-actin, rhodamine phalloidin [red] and PKH67-labeled CD40/IL-4 CLL-EVs [green]) showing internalization of CLL-EVs within CD4+ T cells by 24 hours. Scale bar, 5 μm.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/128/4/10.1182_blood-2015-11-682377/4/m_542f3.jpeg?Expires=1767841020&Signature=XQf7RyiwvhVrreTeSIuWWkWr-4ey2U7bhETQN5rWE9eDogH2A-gbP4u6KizDeKbpszPsd0HOy5779ckobEdmMYXvfJCHpdoacgxWWAWXp97Mdg28Xv4b3vn8LCYblRmK61lbr9lJmcGiE1BzmzkZyVxVnKATx1br6IqK8O0XtC0utlE9pEjooiS0ICrzhxUnyrCABWpzZQXgO31w4B1XILZRyuVGjNZR1jie3TOfEgOx8-7Ttswbtx-5FfwJ5vtMagqocA2TCB5BrcBMMLsVED4ygElTsvGZplN9bfsH22bkqELqrmpvaa6nPjhVu1rrhm7TL9CRCTMrG~YSpVCiAw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 4. CD4+ T cells exposed to autologous EVs from CD40/IL-4–stimulated CLL cells exhibit enhanced immunological synapse signaling and proliferation. (A) Confocal images of T-cell F-actin immune synapse formation (F-actin rhodamine phalloidin, red) interactions with autologous tumor CLL cells (CMAC-labeled, blue) from a representative patient. CD4+ T cells (negatively selected) were treated with purified CLL-EVs for 48 hours (CLL-EVs following CD40/IL-4 stimulation [CD40/IL-4 CLL-EVs]) compared with control EVs from unstimulated CLL cells (CLL-EVs). Treated T cells were then conjugated with autologous CLL B cells (pulsed with superantigen, sAg cocktail and labeled with CellTracker blue dye, CMAC). T-cell conjugates were analyzed by immunofluorescence and confocal microscopy. Confocal images (n = 10 per patient treatment sample) were acquired using an A1R confocal microscope (Nikon) and analyzed with NIS-Elements software (Nikon). Original magnification, ×60. Scale bar, 5 μm. Quantitative image analysis measured the total area (μm2) of F-actin polymerization (F-actin rhodamine phalloidin, red) at CD4+ T-cell contact sites and synapses with CLL cells within the experimental T-cell population. The dot plot shows the mean synapse/interaction area (μm2) ± SEM from a representative patient (from 6 independent CLL samples) (Mann-Whitney t test, **P < .01). (B) Intercellular CD4+ T cell: tumor CLL conjugates formed between CD40/IL-4 CLL-EVs or CLL-EVs treated CD4+ T cells and sAg-pulsed autologous CLL tumor cells were scored using a confocal microscope (scale bar, 5 μm) and image analysis software. Bar chart shows the mean percentage CD4+ T-cell:tumor CLL conjugates ± SEM from 6 independent CLL samples with 100 random T cells analyzed per experiment (Mann-Whitney t test, *P < .05). (C) CD4+ T-cell:tumor CLL conjugates formed during 20 minutes were fixed, permeabilized, and stained with anti-Lck (green). Tumor cells were labeled with CellTracker blue dye, CMAC. T cells are denoted by white dashed lines. Quantitative image analysis (relative recruitment index [RRI]) of Lck accumulation at the immunological synapse is shown, representative evaluation of 20 conjugates from 3 independent experiments, with the mean value shown as a blue bar ± SEM. Scale bar, 5 μm. (D) MFI was calculated for CD4+ T-cell CD69 expression levels (Alexa 488, green fluorescent channel) following treatment with CD40/IL-4 CLL-EVs or CLL-EVs T cells are denoted by white dashed lines. Tumor cells were labeled with CellTracker blue dye, CMAC. Dot plot chart shows the CD69 MFI ± SEM for a representative patient from 6 CLL samples (Mann-Whitney t test, **P < .01). Original magnification, ×60. Scale bar, 5 μm. (E) Nuclear Ki-67 expression, a marker for cellular proliferation, was measured in autologous patient CD4+ T cells exposed to CLL-EVs (CD40/IL-4 CLL-EVs or CLL-EVs) and with and without anti-CD3/-CD28 crosslinking stimulation for 72 hours (-CD3/-CD28 and unstim., respectively). Bar chart shows mean Ki-67 ± SEM for 3 independent CLL patient experiments (1-way ANOVA, **P < .01). (F) The confocal images show increased nuclear (DAPI, blue) Ki-67 expression (Alexa 647 denoted using green with analysis software) in CD4+ T cells (F-actin, red) treated with CD40/IL-4 CLL-EVs compared with coculture with control CLL-EVs. Scale bar, 5 μm. (G) Z-stack confocal analysis (3-dimensional central T-cell region analysis) reveals increased nuclear (DAPI, blue) Ki-67 expression (Alexa 647 denoted using green with analysis software) in CD4+ T cells (F-actin, red) treated with CD40/IL-4 CLL-EVs compared with control CLL-EVs. Scale bar, 5 μm. ANOVA, analysis of variance; DAPI, 4′,6-diamidino-2-phenylindole.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/128/4/10.1182_blood-2015-11-682377/4/m_542f4.jpeg?Expires=1767841020&Signature=T77-SAzAG78u7bxul9ZcS8J-L5Yff7PJLs5HhcawRf9lBiQrK7ztExb9jq53TJK3dKvOcyOk82L6igkd157sREu5S9p~W6L3wIUYIgsjGkWNHKzhEnHE9gDnFwt-Y64hqorPwb4yz-si2xetAyi58FHQgUdGYHeYjBBjTwSBw9hr360bYuexwcPjzGeFGP4WbQNIDNNeFoHg8~BmAt0W64H9v4IDHqTxMqs8ILVyZ3G2PNJkmPH-WpLlzcWNE8AAT8pgx1-1Nr-xSjuLAzQ0WA7597pbxl7DlO-hM74HIktZHwrILxIRFnWJbuh5TN6QNavdE7aTQOCd6aXsWaWSdw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 5. CD4+ T cells exposed to autologous EVs from CD40/IL-4–stimulated CLL cells exhibit enhanced CCL19-induced migration. Primary patient CD4+ T cells were treated with autologous CLL-EVs for 48 hours (CLL-EVs following CD40/IL-4 stimulation [CD40/IL-4 CLL-EVs], compared with control EVs from unstimulated CLL cells [CLL-EVs]). Treated T cells were then allowed to adhere to immobilized ICAM-1 for 10 minutes following exposure to the chemokine CCL19. Video microscopy (Biostation IM; Nikon) was used to observe the migration of CD4+ T cells for 20 minutes before the migration of individual cells was tracked (NIS-Elements analysis software; Nikon). (A) Representative phase-contrast images (original magnification, ×20) and motility tracks (colored) of CD4+ T cells from CLL patients treated with experimental CLL-EVs as denoted (top microscopy images). Representative migratory tracks of individual T cells from CLL patients treated with CLL-EVs are shown in the bottom data plots (n = 20 cells per experiment). Dot plot charts show the (B) speed of migration (micromolar per minute) and (C) distance of migration (μm) for a representative patient from 6 independent CLL samples (Mann-Whitney t test, **P < .01).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/128/4/10.1182_blood-2015-11-682377/4/m_542f5.jpeg?Expires=1767841020&Signature=xtk4isQiVwS4aD7iAth8hh6dVjfR3GTGUMK37PvaCh55mOgzhb1mTMQ9oILR8pkI4iqcOFfZx3r98FJVpB6udVaDow3htcgEKJ7UU891q9WHj2CINYD4Fn1fUIwbBrxgL~q0GFvF-F~Om1Miia8GnFDSfgl0DC1Czc94KdSeqXx7u9LMkRG6gkvMX7Pyt4C4sAVjMK-580ua0ro0IgxH0xnfZHYMoXCzD47zi3IIyOqpFl-cIqcvwdQDG7FIvFyGrq7ljqrvV8pZP4RSmjMmaeUbhaO0TsWlPBTWDSuUFtcJSgZ27nkdwRD0veamclUQ4HVujnP0ZLSPqvZW4RZzYA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 6. Knockdown of miR-363 in parental CLL cells prevents the ability of released CLL-EVs to confer altered functional properties to autologous CD4+ T cells. miR-363 was knocked down in CLL cells using LNA-based antisense oligonucleotides, prior to CD40/IL-4 stimulation and isolation of released EVs (anti-miR-363 compared with scrambled control [Con.] antisense miR oligonucleotides). CD4+ T cells (negatively selected) were treated with purified CLL-EVs from the transfected autologous parental CLL cells. Treated T cells were then conjugated with autologous CLL B cells (pulsed with superantigen [sAg] cocktail and labeled with CellTracker blue dye, CMAC). T-cell conjugates were analyzed by immunofluorescence and confocal microscopy as described in Figure 4. Confocal images (n = 10 per patient treatment sample) were acquired using an A1R confocal microscope (Nikon) and analyzed with NIS-Elements software (Nikon). Original magnification, ×60. Scale bar, 5 μm. Dot plot charts show (A) the mean F-actin polymerization (rhodamine phalloidin, red) synapse/interaction area (μm2) ± SEM from a representative patient (from 3 independent CLL samples), (B) the mean RRI of Lck accumulation (green) at the immunological synapse, representative analysis (±SEM) from 3 independent patient experiments and (C) the CD69 MFI ± SEM for a representative patient from 3 independent CLL patient samples. CD69 expression is denoted in white. Treatment of autologous CD4+ T cells with CLL-EVs derived from miR-363 knocked down cells (anti-miR-363) blocked modulation of T-cell migration rates when compared with EVs derived from control transfected tumor cells (Scrambled Con. miR). Dot plot charts show the (D) speed of migration (micromolar per minute) and (E) distance of migration (μm) for a representative patient from 3 independent CLL samples (Mann-Whitney t test, *P < .05, **P < .01).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/128/4/10.1182_blood-2015-11-682377/4/m_542f6.jpeg?Expires=1767841020&Signature=qbB4~0FqkFh9jxv6K8tXP39KgfLP1UDqvRNoOKwSPJ-k-MbSxw37lqDv01o-mmB7wHwj7BMUfDm9KCaUeZbXWyRIpG46JTVnB55s5D~uPAWyPXHNiswQ5bs3aIIxnIQq36nEfkGnXqX8VLRQgrWpqjJlzu5lau3kPqYqgvo7zxRVOFkXemTAl2oVfbanhxC1kCC~YDdDilBGuoYl049L18caTTcUJe1X5Rjf12FOx5-HoqtyOt9d8yF7hfjToHhGKW2jkAlcdUwnP0688IO4WDMQrEvCRPB0AIWprY4wdh8PhEyXHS9KHarMv~G4iaSrTLAMBS1eouk9YkBMUeLBLw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)