Key Points
The type III TGF-β receptor is a marker that distinguishes “early” and “late” BFU-Es.
TGF-β inhibitors increase early BFU-E cell self-renewal and total erythroblast production.
Abstract
Burst-forming unit erythroid progenitors (BFU-Es) are so named based on their ability to generate in methylcellulose culture large colonies of erythroid cells that consist of “bursts” of smaller erythroid colonies derived from the later colony-forming unit erythroid progenitor erythropoietin (Epo)–dependent progenitors. “Early” BFU-E cells forming large BFU-E colonies presumably have higher capacities for self-renewal than do “late” BFU-Es forming small colonies, but the mechanism underlying this heterogeneity remains unknown. We show that the type III transforming growth factor β (TGF-β) receptor (TβRIII) is a marker that distinguishes early and late BFU-Es. Transient elevation of TβRIII expression promotes TGF-β signaling during the early BFU-E to late BFU-E transition. Blocking TGF-β signaling using a receptor kinase inhibitor increases early BFU-E cell self-renewal and total erythroblast production, suggesting the usefulness of this type of drug in treating Epo-unresponsive anemias.
Introduction
The production of red blood cells in mammals is tightly regulated by erythropoietin (Epo), which stimulates erythropoiesis by promoting survival, proliferation, and terminal differentiation of the colony-forming unit erythroid progenitors (CFU-Es).1,2 Several acute and chronic anemias, including hemolysis, severe trauma-induced anemia, and genetic bone marrow failure disorders such as Diamond-Blackfan anemia (DBA), are not treatable with Epo, because the CFU-Es that respond to Epo are either too few in number or are not sensitive enough to Epo to maintain adequate red blood cell production. Treatment of Epo-resistant anemias requires a drug that acts earlier than Epo in erythropoiesis and that enhances the formation of CFU-Es. One attractive approach is to devise strategies to promote self-renewal of the upstream burst-forming unit erythroid progenitors (BFU-Es).
BFU-Es are the earliest committed erythroid progenitors and are defined based on their ability to produce in methylcellulose-based cultures large colonies of erythroid cells that consist of “bursts” of smaller CFU-E colonies. BFU-Es have limited self-renewal capacity. “Early” BFU-Es, which generate thousands of erythroblasts, presumably have higher self-renewal capacity than “late” BFU-Es that form small colonies. However, environmental factors and cellular pathways contributing to the formation of early and late BFU-Es are largely unknown.
We purified a BFU-E population from mouse fetal livers that when plated in methylcellulose generates ∼62% large and ∼38% small erythroid bursts; presumably, these are “early” and “late” BFU-Es, respectively.3 While recognizing that this BFU-E population was heterogeneous, it was used to great effect in many subsequent experiments.3,4 Given that our BFU-E population is heterogeneous and likely contains multiple subpopulations with different self-renewal capacities, we used single-cell RNA-sequencing technology to dissect the population heterogeneity5,6 and uncovered the type III transforming growth factor β (TGF-β) receptor (TβRIII) as a marker to distinguish early and late BFU-Es. Furthermore, TGF-β inhibitors can increase erythroid cell production by promoting BFU-E self-renewal and therefore might be used to treat Epo-unresponsive anemias.
Study design
We conducted single-cell transcriptome analysis on BFU-Es isolated from mouse fetal livers as previously described.3 Principle-component analysis (PCA) identified 2 BFU-E subpopulations and potential cell-surface markers specific for these subpopulations. TβRIII was identified as a marker to distinguish early and late BFU-Es. Therefore, we purified TβRIII10%lo and TβRIII10%hi BFU-Es for further analyses. For details and additional methods, see supplemental Methods, available on the Blood Web site.
Results and discussion
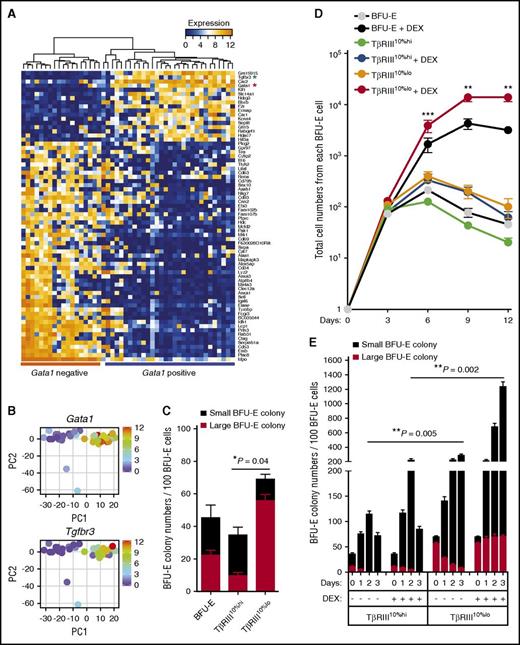
We used single-cell RNA-sequencing technology to dissect the BFU-E population heterogeneity.5,6 After quality control, we obtained high-quality RNA-sequencing results from 48 mouse BFU-E cells (Figure 1A; supplemental Figure 1A). Single-cell expression profiling showed significant variances in gene expression in individual BFU-E cells, including the Gata1, Nr3c1 (the gene encoding glucocorticoid receptor), and Hif1α genes, while the expression of ribosomal protein gene Rps19 and many other genes is more homogeneous (supplemental Figure 1B-C). PCA of single-cell transcriptomes showed that there are 2 principal populations of mouse BFU-E cells (supplemental Figure 1D); the expression of Gata1 is high in one and low in the other (Figure 1A-B; supplemental Table 1). As Gata1 is induced during the BFU-E to CFU-E transition,7 we hypothesized that the Gata1lo population might be enriched in early BFU-Es. We noted that the expression of Tgfbr3, the gene encoding TβRIII, is highly correlated with that of Gata1 (Figure 1A-B). The type III TGF-β receptor, also named betaglycan, does not have a cytoplasmic kinase domain, in contrast to the signaling TβRI and TβRII receptors that phosphorylate and activate downstream SMAD2 or SMAD3 transcription factors.8-10 TβRIII binds all 3 TGF-β isoforms and often forms heterodimers with TβRII, enhancing TGF-β–mediated signaling. Interestingly, definitive erythropoiesis in embryos is disrupted in TβRIII knockout mice, although this might be secondary to liver pathology.11
Single-cell analysis of mouse BFU-E cells identifies TβRIII as a marker that differentiates early and late BFU-E progenitor cells. (A) Unbiased hierarchical clustering of the 96 genes identified by PCA as best separating the transcriptome of the 48 mouse BFU-E cells. Red asterisk, Gata1; green asterisk, Tgfbr3. (B) Gene expression of Gata1 and Tgfbr3 in each single cell is plotted in the PCA analysis. PC, principal component. (C) Colony-forming assays on unfractionated, TβRIII10%hi and TβRIII10%lo BFU-E populations. Large BFU-E colonies have >12 clusters, and small BFU-Es have 5 to 12 clusters as described (*P < .05.). (D) Total BFU-E (c-Kit+CD7110% lo) cells were separated by flow cytometry, and the TβRIII10%hi and TβRIII10%lo populations were subsequently isolated by FACS. Purified TβRIII10%hi or TβRIII10%lo cells were seeded in SFELE medium in the presence or absence of 100 nM DEX. The production of erythroblasts from total BFU-E, purified TβRIII10%hi, and TβRIII10%lo BFU-E cells was quantified over time. Error bars represent mean ± standard deviation from 3 biological replicates (**P < .01; ***P < .001). (E) Colony-forming assays were conducted to determine BFU-E colony numbers from 100 TβRIII10%hi or TβRIII10%lo BFU-E cells cultured under the indicated conditions. Colony-forming assays were performed at 24-hour intervals (**P < .01).
Single-cell analysis of mouse BFU-E cells identifies TβRIII as a marker that differentiates early and late BFU-E progenitor cells. (A) Unbiased hierarchical clustering of the 96 genes identified by PCA as best separating the transcriptome of the 48 mouse BFU-E cells. Red asterisk, Gata1; green asterisk, Tgfbr3. (B) Gene expression of Gata1 and Tgfbr3 in each single cell is plotted in the PCA analysis. PC, principal component. (C) Colony-forming assays on unfractionated, TβRIII10%hi and TβRIII10%lo BFU-E populations. Large BFU-E colonies have >12 clusters, and small BFU-Es have 5 to 12 clusters as described (*P < .05.). (D) Total BFU-E (c-Kit+CD7110% lo) cells were separated by flow cytometry, and the TβRIII10%hi and TβRIII10%lo populations were subsequently isolated by FACS. Purified TβRIII10%hi or TβRIII10%lo cells were seeded in SFELE medium in the presence or absence of 100 nM DEX. The production of erythroblasts from total BFU-E, purified TβRIII10%hi, and TβRIII10%lo BFU-E cells was quantified over time. Error bars represent mean ± standard deviation from 3 biological replicates (**P < .01; ***P < .001). (E) Colony-forming assays were conducted to determine BFU-E colony numbers from 100 TβRIII10%hi or TβRIII10%lo BFU-E cells cultured under the indicated conditions. Colony-forming assays were performed at 24-hour intervals (**P < .01).
We hypothesized that TβRIII could be used as a marker to define BFU-E subpopulations. Indeed, the mouse fetal liver BFU-E cell population expressing the 10% lowest amount of TβRIII (TβRIII10%lo population) is enriched in the percentage of colony-forming BFU-E cells (∼80% pure); 80% form large, early BFU-E colonies that have >12 clusters (supplemental Figure 1E and Figure 1C). Conversely the BFU-E subpopulation with the 10% highest amount of TβRIII (TβRIII10%hi population) is enriched in late BFU-Es. Importantly, the TβRIII10%lo BFU-E population is significantly more responsive to dexamethasone (DEX), which induces self-renewal, than the total BFU-E and TβRIII10%hi BFU-E populations in both erythroid proliferation and colony-formation assays (Figure 1D-E). Thus the TβRIII10%lo mouse fetal liver BFU-E cells represents early BFU-Es that are maximally able to undergo self-renewal divisions when stimulated by corticosteroids and to produce the maximum number of erythroblasts.
Gene set enrichment analysis of the transcriptomes showed that genes expressed in more differentiated hematopoietic cells are more enriched in TβRIII10%hi BFU-E cells than in TβRIII10%lo BFU-E cells (supplemental Figure 1F). In contrast, expression of genes targeted by GATA2, a transcription factor important in hematopoietic stem and progenitor cell self-renewal that is downregulated during the BFU-E to CFU-E transition,2,12 is enriched in TβRIII10%lo compared with TβRIII10%hi BFU-E cells. This result further indicates that TβRIII10%lo BFU-E cells are predominantly early BFU-E cells that have higher self-renewal capacities. Taken together, our data suggest that TβRIII is a marker that distinguishes early from late mouse BFU-E cells.
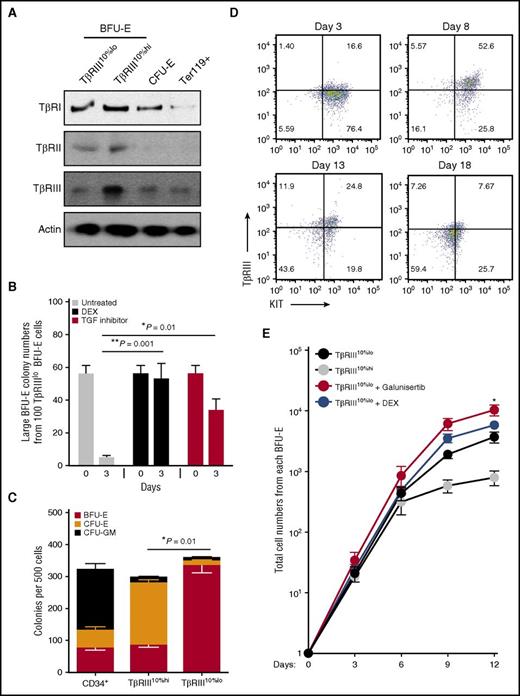
Although some effects of TGF-β signaling on hematopoiesis (erythropoiesis specifically) have been observed,13-15 the mechanism of action of TGF-β on erythroid progenitors remains unclear. We next examined TGF-β signaling.16 SMAD2 and SMAD3 phosphorylation is higher in TβRIII10%hi BFU-E cells than in TβRIII10%lo BFU-E cells; phosphorylated SMAD2 levels are low in CFU-E and undetectable in Ter119+ mouse erythroblasts (supplemental Figure 2A; data not shown). Of the 3 TGF-β receptors, only the expression of TβRIII is upregulated in TβRIII10%hi BFU-E cells relative to the early TβRIII10%lo population (Figure 2A), suggesting that transient upregulation of TGF-β signaling during the early-to-late BFU-E transition is mediated by the increased expression of TβRIII.
TGF-β signaling is transiently upregulated through the transition from early to late BFU-Es. (A) Expression of 3 types of TGF receptors in purified TβRIII10%hi and TβRIII10%lo BFU-E, CFU-E, and Ter119+ cells from mouse fetal livers. (B) Colony-forming assays were conducted to determine large BFU-E colony numbers from 100 TβRIII10%lo BFU-E cells cultured under the indicated conditions. Colony-forming assays were performed at indicated time points. Only large BFU-E colonies with >12 clusters were counted (**P < .01; ***P < .001). (C) Colony-forming assays of BFU-E, CFU-E, and CFU–granulocyte, erythrocyte, monocyte, megakaryocyte in indicated FACS-isolated populations from human CD34+ cord blood cells, based on TβRIII expression as described in “Materials and methods” (*P < .05.). (D) Dynamics of TβRIII expression on cell surface as determined by flow cytometry. Human TβRIII10%lo BFU-E cells were isolated and cultured in medium supplemented with stem cell factor, Epo, interleukin 3, interleukin 6, and DEX during the first 8 days before switching to differentiation medium. (E) Purified human TβRIII10%hi or TβRIII10%lo CD34+ cells were cultured in the presence or absence of 100 nM DEX or 100 nM galunisertib. Galunisertib treatment of significantly increased erythroblast production compared with the TβRIII10%lo CD34+-untreated group. The production of erythroblasts was quantified over time. Error bars represent mean ± standard deviation from 3 biological replicates (*P < .05).
TGF-β signaling is transiently upregulated through the transition from early to late BFU-Es. (A) Expression of 3 types of TGF receptors in purified TβRIII10%hi and TβRIII10%lo BFU-E, CFU-E, and Ter119+ cells from mouse fetal livers. (B) Colony-forming assays were conducted to determine large BFU-E colony numbers from 100 TβRIII10%lo BFU-E cells cultured under the indicated conditions. Colony-forming assays were performed at indicated time points. Only large BFU-E colonies with >12 clusters were counted (**P < .01; ***P < .001). (C) Colony-forming assays of BFU-E, CFU-E, and CFU–granulocyte, erythrocyte, monocyte, megakaryocyte in indicated FACS-isolated populations from human CD34+ cord blood cells, based on TβRIII expression as described in “Materials and methods” (*P < .05.). (D) Dynamics of TβRIII expression on cell surface as determined by flow cytometry. Human TβRIII10%lo BFU-E cells were isolated and cultured in medium supplemented with stem cell factor, Epo, interleukin 3, interleukin 6, and DEX during the first 8 days before switching to differentiation medium. (E) Purified human TβRIII10%hi or TβRIII10%lo CD34+ cells were cultured in the presence or absence of 100 nM DEX or 100 nM galunisertib. Galunisertib treatment of significantly increased erythroblast production compared with the TβRIII10%lo CD34+-untreated group. The production of erythroblasts was quantified over time. Error bars represent mean ± standard deviation from 3 biological replicates (*P < .05).
To test whether TGF-β signaling regulates early BFU-E self-renewal or differentiation, we treated mouse TβRIII10%lo BFU-E cells with galunisertib, a small-molecule inhibitor of the TβRI kinase.17 Colony-forming assays showed that both DEX and galunisertib maintain the number of early BFU-E cells in culture (implying enhancement of BFU-E self-renewal) and increase total mouse red cell production (Figure 2B; supplemental Figure 2B). In the presence of TGF-β, knocking down Tgfbr3 also increases the number of mouse BFU-E colonies, suggesting a specific function of TβRIII in TGF signaling in BFU-E cells (supplemental Figure 2C-D). In contrast, galunisertib had little effect on TβRIII10%hi late BFU-Es or CFU-Es (supplemental Figure 2E-F), indicating a specific function of TGF-β signaling in early BFU-Es. Upregulation of the Hbb and Gata1 genes, which normally occurs during the BFU-E to CFU-E transition, is significantly reduced by both DEX and galunisertib (supplemental Figure 2G). In contrast, expression of Kit, the gene encoding the receptor for stem cell factor that is essential for stem and progenitor cell self-renewal, is upregulated by both DEX and galunisertib. Taken together, these data suggest that blocking TGF-β signaling in early TβRIII10%lo BFU-E cells increases BFU-E cell self-renewal and enhances red cell production.
Human cord blood BFU-Es and CFU-Es can be purified based on differential expression of a group of markers, including CD123 (interleukin 3 receptor) and CD71.18 We found that after a 6-day culture, cord blood CD34+ cells, cells express different levels of TβRIII and CD71 (supplemental Figure 2H). We then determined that CD34+CD123−CD71loTβRIII10%lo cells are highly enriched (70% to 80%) in human BFU-Es (Figure 2C). In contrast, CD34+CD123−CD71loTβRIII10%hi cells are mostly CFU-Es (Figure 2C). Repeating this culture system and purification protocol using adult CD34+ cells generated BFU-Es of the same purity as depicted in Figure 2C, but fewer of them were generated from each adult CD34+ cell than from cord blood CD34+ cells (data not shown), reinforcing the notion that both adult and cord blood CD34+ cells are heterogeneous. We then cultured cord blood CD34+CD123−CD71loTβRIII10%lo BFU-E cells in a 18-day erythroid differentiation system; each BFU-E cell added to the culture generated ∼5000 erythroblasts, of which ∼50% underwent enucleation, as determined by fluorescence-activated cell sorting (FACS) assays and morphological analysis as previously described.4 Expression of TβRIII peaked by day 8 and was gradually downregulated thereafter (Figure 2D). Addition of TGF-β inhibitors significantly increased erythroblast production from human CD34+CD123−CD71loTβRIII10%lo cells (Figure 2E; supplemental Table 2), but not from CD34+CD123−CD71loTβRIII10%hi cells, indicating that TGF-β inhibitors specifically enhance self-renewal of human and murine BFU-E cells. We also observed that some TGF inhibitors show more cytotoxicity in BFU-E cells than others at the concentration we tested (supplemental Table 2).
Consistent with enhancement of BFU-E self-renewal, DEX increases KIT expression in murine BFU-E cells.3,4 Both DEX and galunisertib increase KIT expression in human BFU-E cells (supplemental Figure 2I). Cells treated with galunisertib also underwent normal terminal erythropoiesis and enucleation, as determined by FACS assays and morphological analysis.4 In contrast, treatment of human BFU-E cells with TGF-β quickly downregulated KIT expression. Taken together, these data show that TβRIII, together with other surface markers, can be used for isolation of early human BFU-E cells and that blocking TGF-β signaling in human BFU-E cells can increase human red cell production, which results from enhanced BFU-E self-renewal.
Several signaling pathways have been recently suggested to increase BFU-E self-renewal, of which glucocorticoids have already been used to stimulate erythropoiesis in DBA patients, although severe side effects limit its use.19-21 Here, we have revealed the heterogeneity in BFU-E cells and discovered that inhibiting TGF-β signaling enhances early BFU-E self-renewal and leads to increased red cell production in a corticosteroid-independent manner. This suggests that targeting the TGF signaling pathway might serve as an alternative to corticosteroids in the treatment of DBA and other bone marrow failure disorders.
The single-cell RNA-sequencing data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE81505).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the Whitehead Institute Flow Cytometry Facility, Genome Technology Core and Bioinformatics and Research Computing Facility, as well as the MIT Koch Institute Flow Cytometry Core. They also thank Arinola O. Lampejo for assistance with short hairpin RNA knockdown experiments.
This study was supported by grants from Defense Advanced Research Projects Agency (#HR0011-14-2-0005) (H.F.L.), Department of Defense/US Army Medical Research and Materiel Command (W81WH-12-1-0449), and National Institutes of Health/National Heart, Lung, and Blood Institute (2 P01 HL032262-25), as well as by research support from the Diamond-Blackfan Anemia Foundation and Diamond Blackfan Anemia Canada. X.G. was supported by a postdoctoral fellowship from the Leukemia and Lymphoma Society. H.-Y.L. was supported by a postdoctoral fellowship from the Charles H. Hood Foundation.
Authorship
Contribution: X.G., H.-Y.L., and H.F.L. designed the experiments; X.G., H.-Y.L., Y.-F.L., D.L., Y.F., P.C., J.E., and R.R.E. performed the experiments; E.L.d.R., H.L., M.I.B., C.Z., and P.C. conducted bioinformatic analyses; X.G., H.-Y.L., E.L.d.R., H.L., G.Q.D., and H.F.L. wrote the manuscript; and all authors discussed the results and commented on the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Harvey F. Lodish, 9 Cambridge Ctr, Cambridge, MA 02142; e-mail: lodish@wi.mit.edu.
References
Author notes
X.G. and H.-Y.L. contributed equally to this study.