Abstract
Survival for childhood acute lymphoblastic leukemia (ALL) now approaches 90% with risk adapted therapy based on National Cancer Institute risk group (NCI RG) at diagnosis, somatic lymphoblast genetics, and early response to therapy as measured by minimal residual disease (MRD). Recent studies have identified multiple somatic genetic mutations in ALL, some of which confer increased risk of relapse or identify opportunities for additional targeted therapies. However, there are few genome sequencing analyses of representative cohorts of childhood ALL treated on contemporary regimens.
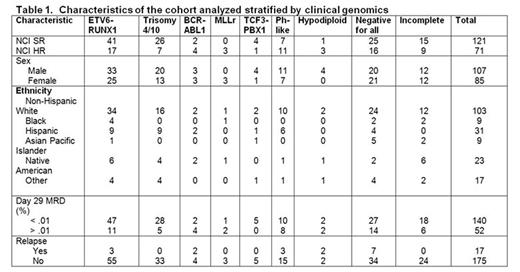
To define the mutational landscape of childhood B-ALL, we performed whole exome sequencing (WES) on diagnostic tumor and remission samples of 192 patients with B-ALL consecutively enrolled between 4/1/06-9/8/06 on the Children's Oncology Group AALL03B1 classification trial that enrolled 11,145 patients up to age 30, which included patients enrolled on clinical trials for standard-risk (SR) B-ALL (N=5226, age 1-10 years and white blood cell count (WBC) < 50,000/uL) and high-risk (HR) B-ALL (N=2907; age ≥10 years or WBC ≥50,000/uL). An 8-gene expression low density array card identified Ph-like patients, and single nucleotide polymorphism arrays and multiplex ligation assays were used to determine DNA copy number alterations. Approximately 2/3 NCI SR and 1/3 NCI HR patients with sufficient banked samples were selected to reflect a population-based cohort (Table 1). Comparison of those selected vs. those not revealed significantly more NCI SR patients with a higher WBC and MRD, and fewer with double trisomy 4 and 10. Selected NCI HR patients were younger, had a higher WBC, and had more ETV6/RUNX1.
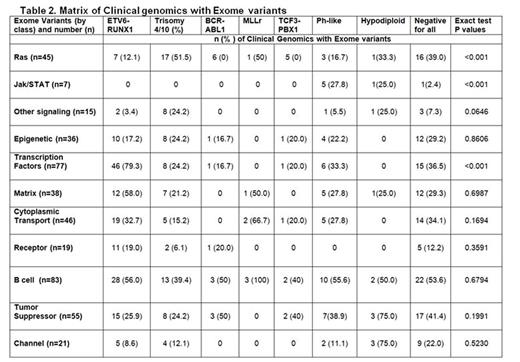
There were a total of 3576 non-silent mutations with a median of 15 mutations/case (range 1-134). One case had 102 non-silent mutations and a germline mutation in MSH3. The additional mutations clustered in 11 pathways (Table 2), several of which were novel including cell-matrix interaction (e.g. SSPO, FAT1) and intracellular trafficking/cytoplasmic transport (e.g. DYNC2H1, ANK3, UNC13C). Most commonly altered were B-cell development (49.4%), transcription factors (45.8%), tumor suppressor genes (32.7%), cytoplasmic transport (27.3%), and Ras signaling (26.7%). Several pathways Ras signaling, Jak/STAT, and transcription factor mutations/deletions were associated with genetic lesions commonly used for risk stratification. Other pathways (epigenetic, B-cell development, or tumor suppressor) occurred in all subtypes. The most commonly identified genetic mutations were NRAS (n=33), KRAS (n=26), FLT3 (n=13), PAX5 (n=10), CREBBP (n=10), XBP1 (n=9), WHSC1 (n=7), and UBA2 (n=7). XBP1 and UBA2 mutations were novel. XBP1 (X-Box binding Protein 1) encodes a transcription factor that regulates the unfolded protein response. UBA2 (ubiquitin like modifier activating enzyme 2) encodes a protein involved in sumoylation to regulate protein structure and intracellular localization. Univariable analysis revealed no significant associations with any of these pathways or mutations with an increased risk of relapse with the exception of IKZF1 mutations or deletions (n=36; p=0.0087). Multivariable analysis modeling including IKZF1, age, presenting WBC, gender, NCI RG, ETV6/RUNX1, BCR/ABL1, Ph-like, white race, and MRD revealed only BCR/ABL1 and MRD positivity being significantly predictive of relapse. However, the risk of relapse was significantly increased based on the number of mutations identified in any one single patient (HR 1.02, 95% CI 1.01-1.023, p < 0.0004). Of note, only 18 patients (NCI SR, n=8, NCI HR, n=10) were Ph-like in this cohort, likely explaining the lack of significance between Ph-like status and an increased risk of relapse in this analysis.
In summary, WES of a consecutively enrolled cohort of NCI SR and HR patients revealed a large number of novel genetic mutations that could be broadly assigned to 11 classes. Outcomes for patients overall were not influenced by any one of these classes, demonstrating that sentinel genetic alterations currently used in risk stratification are of paramount importance in directing therapy intensification for ALL. These data provide important information about pathways commonly mutated in childhood ALL, identifying classes of drugs that can be considered for clinical testing to further improve outcome.
Loh:Bristol Myers Squibb: Membership on an entity's Board of Directors or advisory committees; Abbvie: Research Funding. Hunger:Amgen: Equity Ownership; Pfizer: Equity Ownership; Merck: Equity Ownership; Jazz Pharmaceuticals: Honoraria; Sigma Tau Pharmaceuticals: Honoraria; Erytech: Honoraria; Patent: Patents & Royalties: Dr. Hunger is a co-inventor of a patent (#8658,964) for the identification of novel subgroups in high risk B-ALL and outcome correlations and diagnostic methods related to the same; Spectrum Pharmaceuticals: Honoraria.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal