Abstract
Background
C/EBPα plays a pivotal role in myeloid differentiation at the CMP to GMP transition point, where it interacts with other transcription factors (TFs) implicated in haematopoiesis. CEBPA mutations are common in acute myeloid leukaemia (AML), predominantly in patients with M1 and M2 French-American-British (FAB) morphological classifications, but relatively little is understood about the pre-leukaemic alterations caused by mutated CEBPA. Murine models have established N321D as a particularly potent CEBPA mutation which causes AML with high mortality (Togami et al, Experimental Hematology, 2015). We aimed to develop an inducible expression system for CEBPA N321D in a cellular model which replicates early haematopoietic progenitors, to study the effects of this mutation on gene expression profiles relevant for malignant haematopoiesis.
Methods
We constructed a Piggy-bac Tet-on inducible expression system which has a 2A peptide mechanism enabling simultaneous expression of both N321D and mCherry fluorescent protein from the same transcript (Fig. 1). We also constructed a control with inducible expression of mCherry. These two plasmids were then transfected into the mouse progenitor cell line Hoxb8-FL (Redeckeet al, Nature Methods, 2013), which is conditionally immortalized and models multipotent myelo-lymphoid progenitors. Single cell clones were established and selected for analysis on the basis of cell growth and mCherry fluorescence on induction. RNA was collected post-induction and without induction at 24, 48 and 72 hours in two replicates each from the N321D clone and from the empty control vector. RNA-seq data was aligned to the mouse genome using STAR aligner, processed to generate high throughput sequencing counts, and finally differential expression analysis was performed between N321D and the control.
Results
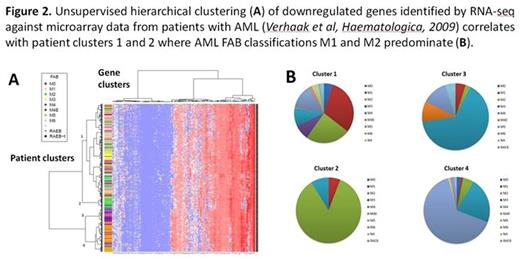
Differential expression analysis identified 172 downregulated and 60 upregulated genes after N321D induction. Further analysis of the 172 downregulated genes against online published datasets of gene expression (Gene Expression Commons, https://gexc.stanford.edu), revealed that 19 of these genes are normally upregulated at the CMP to GMP transition. These include genes such as Hck, Met, Hdac8 and Kdm7a which have been previously implicated in haematological malignancy and which may provide novel insights into the leukaemic process fostered by the CEBPA N321D mutation. To further validate our data, we performed unsupervised hierarchical clustering of previously published microarray data from a large collection of over 400 AML expression profiles (Verhaaket al, Haematologica, 2009) using the genes identified in our study, and found that patient samples who had predominantly FAB classifications M1 and M2 clustered together (Fig. 2A,B), as would be expected in CEBPA-mutated AML.
Conclusions
Our inducible expression system has the potential to provide novel insights into altered gene expression caused by induction of mutated CEBPA. In particular, our cellular model replicates an early stage of haematopoiesis, and implicates genes which were not previously known to interact with CEBPA. The importance of these genes in CEBPA N321D-mediated re-configuration of the myeloid transcriptional regulatory network requires further analysis.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal