Abstract
High-risk Multiple Myeloma (MM) is characterized by unresponsiveness to multiple therapies, rapid disease progression and short overall survival, and may be significantly different from relapsed MM, where aggressiveness is usually a result of drug-resistance associated to clonal selection. Several gene expression-based signatures have been proposed in the past years, however the identification of high-risk patients at diagnosis still represents a challenge. Next generation high-throughput sequencing technologies have enabled a deeper insight into cancer genomes and transcriptomes at an unprecedented level of detail. MMRF CoMMpass is a longitudinal, prospective observational study, started in 2011, that aims to collect and analyze sequencing and clinical data from >1,000 MM patients at initial diagnosis and at relapse. CoMMpass is a real world observational study and, as such, reflects the therapeutic heterogeneity seen across patient populations and provides a unique opportunity to correlate molecular profiles, genomic alterations and clinical characteristics of MM with treatment outcome.
Here we present a network approach to identify high-risk myeloma patients developed using next generation sequencing data from 450 patients in the IA7 release of CoMMpass. We generated MMNet, an integrated network model of newly diagnosed myeloma based on RNA-seq, Whole-Exome (WXS) and Whole-Genome (WGS) data correlated with clinical outcomes. MMNet consisted of 37 modules of coexpressed genes, that were further characterized by functional enrichment analysis and correlation with clinical traits and genomic alterations, i.e. somatic mutations and copy number alterations inferred from WGS and WXS data.
A total of 89 progression/death events have been reported for the cohort within the second year since the beginning of the study. Cox regression analysis identified a module of co-expressed genes whose over-expression was significantly correlated with early relapse (<2yr) (HR 1.75, 95%CI = 1.169-2.614, p=0.005). The module was also associated to stage III R-ISS, high clonality (>4 clones) and high mutational burden, as well as higher percentage of plasma cells in both bone marrow and peripheral blood, which are traits associated with high-risk disease. Module expression was also up regulated in patients with mutations in TP53 and MAX, 13q deletion and 1q amplification.
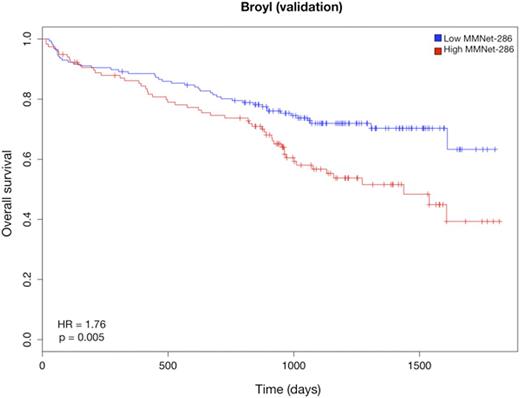
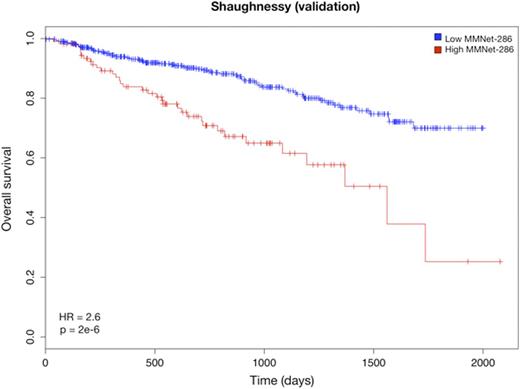
We further narrowed down the signature to 286 genes (the MMNet-286 signature) strongly correlated with time to Event Free Survival (EFS) (r = -0.81, p = 0). This gene-set was significantly enriched for several pathways including Cell Cycle, DNA repair and Homologous Recombination (q < 0.01). Cox regression analysis showed that the two clusters induced by MMNet-286 discriminated between lower and higher risk patients with respect to EFS (HR = 2.22, 95% CI = 1.505-3.295, p = 4.007e-5) (Fig. 1). The prognostic value of MMNet-286 was confirmed on two independent datasets: Broyl-2010 (HR = 1.76, 95% CI = 1.182-2.642, p = 0.005) and Shaughnessy-2006 (HR = 2.65, 95% CI = 1.746-4.031, p = 2.03e-6) (Fig. 2 and 3). The Broyl-2010 dataset consisted of 275 samples from newly diagnosed myeloma patients included in the HOVON65/GMMG-HD4 trial (GSE19784). The Shaughnessy-2006 dataset consisted of 559 samples from newly diagnosed patients pre-TT2 and -TT3 treatments (GSE2658). Comparison of MMNet-286 with previous high risk signatures and disease classes revealed an overlap of five genes with the UAMS-70 signature, twelve genes with the EMC-92 signature and fifteen genes with the set of up-regulated genes in the UAMS PR class, for which the coexpression module was enriched.
In Conclusion, our results demonstrate the advantages of employing integrated network models to identify prognostic features based on next generation sequencing data from large cohort of patients. Applications of the MMNet-286 signature include the generation of a prognostic assay (i.e. NanoString) for the identification of high-risk patients. Future work will aim at validation of the signature in larger cohorts from CoMMpass and other studies.
Kaplan-Meier curves of event free survival in the MMRF cohort stratified by the MMNet-286 signature.
Kaplan-Meier curves of event free survival in the MMRF cohort stratified by the MMNet-286 signature.
Kaplan-Meier curves of overall survival in the Broyl cohort stratified by the MMNet-286 signature.
Kaplan-Meier curves of overall survival in the Broyl cohort stratified by the MMNet-286 signature.
Kaplan-Meier curves of overall survival in the Shaughnessy cohort stratified by the MMNet-286 signature.
Kaplan-Meier curves of overall survival in the Shaughnessy cohort stratified by the MMNet-286 signature.
Chari:Novartis: Consultancy, Research Funding; Array Biopharma: Consultancy, Research Funding; Pharmacyclics: Research Funding; Amgen Inc.: Honoraria, Research Funding; Celgene: Consultancy, Research Funding; Janssen: Consultancy, Research Funding; Takeda: Consultancy, Research Funding. Cho:Genentech Roche: Membership on an entity's Board of Directors or advisory committees, Research Funding; Bristol-Myers Squibb: Membership on an entity's Board of Directors or advisory committees, Research Funding; Agenus, Inc.: Research Funding; Ludwig Institute for Cancer Research: Membership on an entity's Board of Directors or advisory committees; Janssen: Consultancy, Research Funding. Barlogie:Signal Genetics: Patents & Royalties. Dudley:GlaxoSmithKline: Consultancy; Janssen Pharmaceuticals, Inc.: Consultancy; Ayasdi, Inc.: Equity Ownership; Ecoeos, Inc.: Equity Ownership; NuMedii, Inc.: Equity Ownership; Ontomics, Inc.: Equity Ownership; AstraZeneca: Speakers Bureau; NuMedii, Inc.: Patents & Royalties; Personalis: Patents & Royalties.
Author notes
Asterisk with author names denotes non-ASH members.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal