Abstract
Background: Patient-derived xenografts (PDXs) are key models for interrogating the biology of tumor cells that poorly survive in vitro. In particular, over the last decade, immunodeficient mouse models have been extensively used to assess the in vivo growth potential of human leukemia, to provide insights into its biology, and to perform preclinical validation of therapies. Still, only a fraction of the cases of acute myeloid leukemia (AML) are able to engraft into mice, and the biological and clinical correlates of the ability to generate PDXs are unknown.
Methods: Primary AML harvested from 52 patients at diagnosis (n=37, 71%), at relapse after treatments (n=15, 29%), or both (n=6) were purified and infused into non-irradiated NOD-SCID γ-chain null (NSG) mice. Upon leukemia engraftment, assessed by multiparametric flow cytometry, mice were sacrificed and leukemic cells were isolated, characterized, and reinfused in serial recipients, in up to four serial passages. Gene expression profile was analyzed using Illumina microarray, and deregulated genes and processes identified by pairwise LIMMA analysis and classified using Gene Ontology (GO) and Gene Set Enrichment Analysis (GSEA) curated databases. The mutational asset of infused AML was assessed through targeted resequencing, using a custom panel comprising 192 targets and based on the Agilent Haloplex HS technology.
Results: Twenty-six out of 52 primary AML samples (50%) generated xenografts. Engraftment and growth kinetics of the human leukemic cells were highly consistent among littermates, and specific for each tested leukemia. Circulating leukemic cells were firstly detected in the peripheral blood of animals at a median time of 22.5 days (range 14 - 150). In vivo growth allowed expansion of infused AMLs in bone marrows and spleens of the animal, with a median fold increase of 3.5 (range 0.1 - 351.4). The gene expression profile of xenografts was reproducible amongst littermates and recapitulated the features of parental AML: genes deregulated in xenografts accounted for 9.1% of the transcript assessed, with substantial overlap in the genes and processes deregulated in each of the studied cases. GO and GSEA demonstrated the selective deregulation of genes involved in cell proliferation (CDC20, AURKA), syster chromatyde organization (CENPF CEP170) and myeloid differentiation (AZU1, MPO, MYADM, CTSG). Of note, the ability to generate xenografts was conserved when AML cells were challenged at different time-points during the clinical history of the patients, with leukemia harvested at relapse after transplantation displaying a more aggressive behavior. Similarly, upon serial transfer AML exhibited an accelerated growth kinetic.
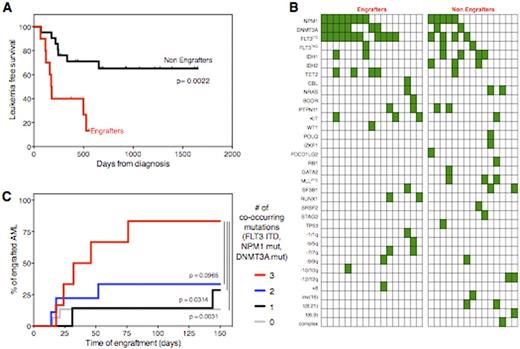
Engraftment in mice significantly correlated with poor patient prognosis: AML engrafters had dramatically lower leukemia free-survival rates compared to non-engrafters (median 5.9 vs. 21.8 months after induction chemotherapy, p=0.0022, Fig. 1A), confirmed also by multivariate analysis (p=0.002).
Also the mutational profile differed greatly between engrafters and non-engrafters, as summarized in Fig. 1B. In particular, while the presence of an aberrant karyotype was not associated with PDX generation, FLT3 internal tandem duplication, DNMT3A and NPM1 mutation were all significantly associated to engraftment (p=0.0244, p=0.009 and p=0.0437 respectively). In particular the co-occurrence of mutations in these three genes, recently reported to confer very poor prognosis to AML patients (Papaemmanuil et al, NEJM 2016), markedly enhanced the ability to generate PDXs (Fig.1C).
Conclusion: These data show that engraftment into immunodeficient mice mirrors the biology of primary human leukemia, providing a proxy to select cases with a higher chance to generate PDXs. Further comparisons between AML capable or not to generate PDXs might provide novel markers of leukemia aggressiveness and rationales for targeted therapies.
Bonini:TxCell: Membership on an entity's Board of Directors or advisory committees; Molmed SpA: Consultancy. Ciceri:MolMed SpA: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal