Abstract
An increasing number of CSF3R cytoplasmic domain truncation (nonsense and frameshift) and missense mutations are being identified with the wide application of next generation sequencing (NGS) to leukemia patient samples. Previous studies have demonstrated oncogenic potential of some CSF3R truncation mutations seen in severe congenital neutropenia, chronic neutrophilia leukemia (CNL), and Philadelphia-negative atypical chronic neutrophilic leukemia (aCML) patients. However, full understanding of the spectrum of CSF3R cytoplasmic mutations harboring leukemogenic potential as well as the associated mechanisms of enhanced signaling remains incomplete. In the current study, we leveraged NGS data to first validate that CSF3R cytoplasmic mutations are prevalent in CNL/aCML/unclassified myeloproliferative neoplasm (8.5%, n=212) patients and to a lesser extent in AML patients (1.3%, n=378). We did not observe CSF3R cytoplasmic mutations in other hematologic malignances. Notably, 72.2% of CSF3R cytoplasmic mutations co-occur with the membrane proximal mutation, CSF3R T618I. To evaluate the leukemogenic potential of these cytoplasmic mutations, we first investigated the minimal mutated elements necessary for CSF3R leukemogenic potential. We identified that truncation mutations of the CSF3R cytoplasmic tail between T738 and Q823 all harbor leukemogenic potential. In contrast, CSF3R truncations before E700X did not respond to G-CSF and did not transform Ba/F3 cells even when combined with T618I. Truncation mutations between E700X and T738X demonstrated reduced G-CSF sensitivity, but could transform Ba/F3 cells when combined with CSF3R T618I. Surprisingly, all the cytoplasmic missense mutations (Q754A, R769H, L777F, T781I, S795R and Q823H), seen in the patient samples did not harbor leukemogenic potential. We further identified that missense mutations disrupting the di-leucine residues L776/777, S772, or ubiquitin binding lysine residues (K655, K785 and K704) demonstrated transforming capacity.
Mechanistically, truncation mutations prior to Q793 are associated with delayed receptor internalization due to the disruption of the internalization motifs (amino acids 772-778 and 779-792), whereas truncation mutations between Q793 and Q823 decrease receptor degradation, which may be associated with loss of the de-phosphorylation domain (amino acids N818-F836). All truncation mutations before Q823 demonstrate altered patterns of molecular weight bands and are sensitive to the Golgi inhibitor brefeldin A, indicating that CSF3R glycosylation plays a role in the leukemogenic potential of these truncations. We also showed that K704 is a potential ubiquitin binding site, and substitution of K704A demonstrated reduced ubiquitination. Downstream, leukemogenic cytoplasmic mutations all induce sustained STAT5 activation and are sensitive to the JAK inhibitor ruxolitinib. In summary, we have defined the region of the CSF3R cytoplasmic domain in which truncating mutations exhibit the capacity for leukemogenesis. This information will be useful for evaluating the potential clinical relevance of CSF3R mutations appearing in genomic reports from patients with myeloid leukemia.
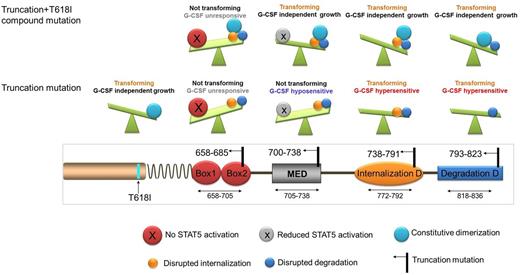
Schematic illustration of CSF3R truncation mutations and the functional consequences. CSF3R truncation mutations at 658-685 which abrogate STAT5 activation do not transform BaF3 cells and do not respond to G-CSF, even when combined with T618I mutation. Truncation mutations at 700-738 do not transform Ba/F3 cells alone, however, these truncations can transform Ba/F3 when combined with T618I and are hyposensitive to G-CSF due to the disruption of the mitogenic enhancing domain. Truncation mutations between 738-791 and 793-823 transform Ba/F3 and induce G-CSF hypersensitivity due to the interruption of the internalization and the de-phosphorylation domain respectively. MED: mitogenic domain; D: Domain.
Schematic illustration of CSF3R truncation mutations and the functional consequences. CSF3R truncation mutations at 658-685 which abrogate STAT5 activation do not transform BaF3 cells and do not respond to G-CSF, even when combined with T618I mutation. Truncation mutations at 700-738 do not transform Ba/F3 cells alone, however, these truncations can transform Ba/F3 when combined with T618I and are hyposensitive to G-CSF due to the disruption of the mitogenic enhancing domain. Truncation mutations between 738-791 and 793-823 transform Ba/F3 and induce G-CSF hypersensitivity due to the interruption of the internalization and the de-phosphorylation domain respectively. MED: mitogenic domain; D: Domain.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal