Abstract
Next-generation sequencing (NGS) has revealed new insight into the complexity of clonal and subclonal architecture of multiple myeloma (MM). The introduction of targeted studies allows the detection of mutations with very low allele frequencies at affordable price. However, there is few information about the prognostic impact of this mutational profile in series of homogeneously treated MM patients.
In this study, we analyzed the most frequently mutated genes in MM from patients included in a randomized clinical trial applying the highest read depth to date. We performed a correlation study between our results and cytogenetics aberrations and clinical outcomes.
We used Ampliseq Library Kit 2.0 to target 77 genes (M3Pv2.0 panel) related to critical pathophysiological pathways, associated to drug resistance or targetable with molecular drugs in MM. We sequenced DNA (15ng) from CD138+ purified plasma cells from 80 MM patients at diagnosis included in GEM2010MAS65 clinical trial. We sequenced at an average read depth of 1500X and using Ion Proton sequencer (Thermo Fisher, USA, PaloAlto, CA). We used Ion Reporter software applying in-house modifications in call variants process. We also sequenced DNA from CD138 negative population in 30% of patients in order to filter out potential artifacts and stablish conditions for excluding germline variants. We first fitted a cox-proportional hazard model to predict survival and a second approach using a penalized regression LASSO.
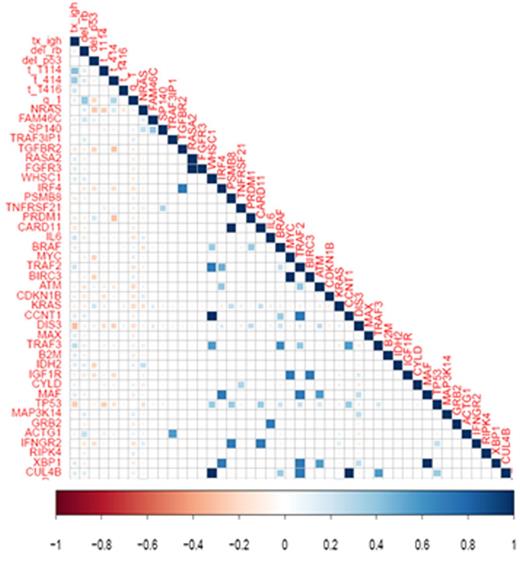
We identified 137 gene mutations in the 80 patients analyzed, 54 mutations were predicted pathogenic by Shif and Poliphen and 65 have been described in COSMIC database. 85% of patients harbored at least 1 mutation. The most frequently mutated genes were KRAS (16%), DIS3 (15%), NRAS (13%), BRAF (6%), and TP53 (5%), accounting for the 54% of the total number of mutations. The most frequently mutated pathways were RAS and NFKB in 34% and 28% of patients respectively. Mutational allele frequency for KRAS, BRAF and TP53 was, in all cases, lower than 50% while DIS3 showed mutations in a broad range (from 2 to 85%). Only one patient presented one mutation in NRAS at 73 % of allele frequency. For patients with more than one mutation, two different scenarios have been found. Some patients showed several genes mutated in a similar allele frequency values. On the contrary, a complex subclonal structure was confirmed in 3 patients, with mutations at very different clonal frequency. CD138 negative fraction was sequenced to confirm that these mutations were exclusive of plasma cells. The first of them had two mutations in DIS3 at 8 and 53% of allele frequency, second in KRAS and DIS3 at 13 and 63% respectively and the third in NRAS and IFNGR2 at 5 and 37% respectively. We found no differences in the survival based on pathway, mutational allele frequency or number of mutations. We also investigated pairwise associations between events and we not found significant correlations (Figure 1). Cox-proportional hazard model only showed a negative impact in survival of patients with TP53 mutations.
This ultra-deep targeted sequencing strategy is able to detect mutations in most of patients, at very low allele frequency and using a minimal amount of DNA. Despite of its huge capacity to detect mutations, we have not identified new prognostic factors in MM patients treated with highly efficient therapies. However, this study again confirms the complexity of the genomic landscape of MM and the great heterogeneity between patients. The role of theses mutations at relapse as well as in the subsequent treatment effectiveness remains unknown and should be preferably investigated in larger cohorts of homogeneously treated MM patients.
Martínez-López:Novartis: Honoraria, Speakers Bureau. Oriol:Janssen: Honoraria, Membership on an entity's Board of Directors or advisory committees; Amgen: Honoraria, Membership on an entity's Board of Directors or advisory committees; Celgene: Honoraria, Membership on an entity's Board of Directors or advisory committees. Stewart:celgene: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal