Abstract
MicroRNAs (miRNAs) are small RNAs that bind Argonaute (Ago) family proteins and recruit them to target mRNAs based on seed sequence complementarity, thereby causing mRNA degradation and/or translational repression. Multiple miRNAs are dynamically expressed during erythropoiesis. Deletion of miR-144/451, the most abundantly expressed erythroid miRNA gene, causes anemia and increased red cell sensitivity to oxidant stress, in part by de-repressing the target mRNA Ywhaz. However, the total number of mRNAs targeted by miR-144/451 is unknown.
To identify erythroblast miR-144/451 target genes comprehensively, we used a technique named HITS-CLIP to sequence mRNA fragments bound to Ago proteins in erythroblasts from miR-144/451 wild-type (WT) and knockout (KO) mouse fetal livers. Using a novel peak-calling algorithm (YODEL), we determined that Ago binds to 6,651 peaks on 3,533 mRNAs (Fig 1A). Of these, 1/3rd (2,212 peaks on 1,414 mRNAs) were depleted in KO, indicating that Ago binding to these sites depended on the presence of miR-144/451. Seed sequences for deleted miRNAs (451, 144-5p, 144-3p) were enriched in the depleted peak set (Fig 1B), while seeds for non-deleted miRNAs (486a, 16-5p, 122-5p) were concentrated in the non-depleted set (Fig 1C), validating the specificity of our analysis.
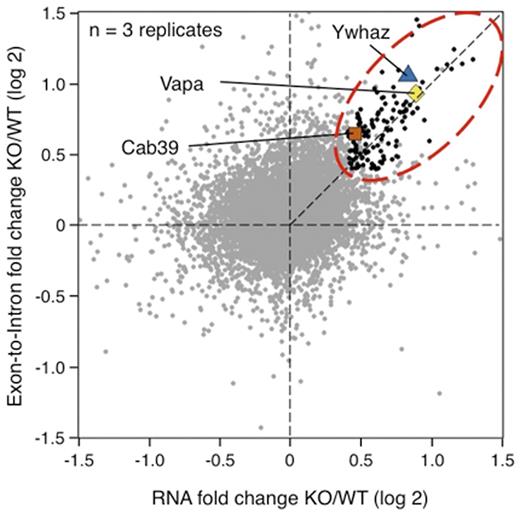
We then performed RNA-Seq and quantitative proteomics by TMT-mass-spectrometry on WT and KO erythroblasts. mRNAs directly targeted by miRNAs are expected to be stabilized in KO, compared to mRNAs altered in abundance through indirect transcriptional effects. We inferred mRNA stability from RNA-Seq data by calculating the ratio of exonic and intronic signals in KO and WT cells (Fig 2). Using these criteria, 131 mRNAs showed increased stability (genes within red dotted oval) in KO erythroblasts compared to WT, 100 of which also showed increased protein levels by mass spectrometry. In contrast, only 12 genes showed reduced RNA stability and protein in KO. Notably, very few genes were altered solely at the protein level, indicating that repression by miR-144/451 occurs largely through mRNA degradation, not translation inhibition.
Surprisingly, most mRNAs bound by miR-144/451-Ago complexes did not show altered stability or translation (1,414 bound vs. 131 altered mRNAs). To investigate this mismatch, we examined HITS-CLIP peaks for additional features predictive of target regulation. Peaks in 3prime UTRs were more likely to regulate mRNA levels than peaks in coding exons (p=10-7), and peaks containing canonical seed sequences matching miR-144/451 were more likely to regulate mRNA levels than those lacking them (p=10-5). This indicates that while miRNAs recruit Ago to a large number of mRNA sites, the location of the binding site within the mRNA, and the degree of seed match, are important determinants of target regulation.
Our combined studies identified numerous mRNAs that are targeted directly by miR-144/451. Ndufb5, Cox10 and Hccs showed increased mRNA stability and increased protein in KO, and showed miR-144/451 dependent HITS-CLIP peaks with canonical seed sequences. All three mRNAs encode components of the mitochondrial electron transfer chain (ETC), or are required for normal ETC function. Preliminary studies show that KO erythroblasts exhibit increased ETC activity consistent with de-repression of its component genes.
Overall, our results demonstrate that miRNA-guided binding of Ago proteins to mRNAs is insufficient to produce mRNA repression, and that additional modifying variables determine whether physical interaction leads to repression. This finding is of general relevance to miRNA biology. Moreover, our studies provide a more comprehensive set of erythroblast miR-144/451 mRNA targets for further study, including components of the mitochondrial electron transfer chain.
(A) Volcano plot showing HITS-CLIP peaks in KO vs. WT erythroblasts. Known targets of miR-144/451 (Ywhaz, Cab39 and Vapa) are indicated. (B) Distribution of canonical seed sequences of miRNAs deleted in KO mice. (C) Distribution of canonical seed sequences of miRNAs not deleted in KO mice.
(A) Volcano plot showing HITS-CLIP peaks in KO vs. WT erythroblasts. Known targets of miR-144/451 (Ywhaz, Cab39 and Vapa) are indicated. (B) Distribution of canonical seed sequences of miRNAs deleted in KO mice. (C) Distribution of canonical seed sequences of miRNAs not deleted in KO mice.
Alteration in gene expression between KO vs WT erythroblasts at the mature mRNA level (X-axis) and the RNA stability level (Y-axis). Known targets of miR-144/451 (Ywhaz, Cab39 and Vapa) are indicated. mRNAs with increased stability and abundance are indicated as black dots within a red dotted oval.
Alteration in gene expression between KO vs WT erythroblasts at the mature mRNA level (X-axis) and the RNA stability level (Y-axis). Known targets of miR-144/451 (Ywhaz, Cab39 and Vapa) are indicated. mRNAs with increased stability and abundance are indicated as black dots within a red dotted oval.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal