Key Points
Inhibition of HSP90 targets multiple dependences in mantle cell lymphoma.
Clinically available HSP90 inhibitors overcome ibrutinib resistance in vitro and in vivo.
Abstract
The Bruton tyrosine kinase (BTK) inhibitor ibrutinib induces responses in 70% of patients with relapsed and refractory mantle cell lymphoma (MCL). Intrinsic resistance can occur through activation of the nonclassical NF-κB pathway and acquired resistance may involve the BTK C481S mutation. Outcomes after ibrutinib failure are dismal, indicating an unmet medical need. We reasoned that newer heat shock protein 90 (HSP90) inhibitors could overcome ibrutinib resistance by targeting multiple oncogenic pathways in MCL. HSP90 inhibition induced the complete degradation of both BTK and IκB kinase α in MCL lines and CD40-dependent B cells, with downstream loss of MAPK and nonclassical NF-κB signaling. A proteome-wide analysis in MCL lines and an MCL patient-derived xenograft identified a restricted set of targets from HSP90 inhibition that were enriched for factors involved in B-cell receptor and JAK/STAT signaling, the nonclassical NF-κB pathway, cell-cycle regulation, and DNA repair. Finally, multiple HSP90 inhibitors potently killed MCL lines in vitro, and the clinical agent AUY922 was active in vivo against both patient-derived and cell-line xenografts. Together, these findings define the HSP90-dependent proteome in MCL. Considering the disappointing clinical activity of HSP90 inhibitors in other contexts, trials in patients with MCL will be essential for defining the efficacy of and mechanisms of resistance after ibrutinib failure.
Introduction
Despite recent improvements in treatment, mantle cell lymphoma (MCL) remains an incurable disease for the majority of patients.1 The molecular hallmark of MCL is aberrant expression of cyclin D1, most commonly through IGH@-CCND1 translocation,2 which drives cell-cycle progression through cyclin-dependent kinases (CDKs) 4 and 6. The majority of MCLs have constitutive activation of the B-cell receptor (BCR) signaling pathway through the kinases Bruton tyrosine kinase (BTK), LYN, SYK, and downstream pathways, including MAPK and classical NF-κB.3,4 Inhibition of BTK with the small-molecule inhibitor ibrutinib is an effective strategy in these MCLs,5 as well as in chronic lymphocytic leukemia (CLL)6 and activated B-cell-like diffuse large B-cell lymphoma.7,8 However, ∼32% of patients with MCL fail to respond to ibrutinib and median progression-free survival is only 14 months.5 Outcomes after ibrutinib failure are dismal due to a lack of effective agents.9 Thus, the most important clinical question facing this field is how to overcome intrinsic and acquired ibrutinib resistance?
Reports of resistance to ibrutinib began emerging soon after its accelerated approval by the US Food and Drug Administration for MCL (in 2013) and CLL (in 2014). Mutations affecting BTK residue C481, which is bound covalently by ibrutinib, were identified in patients with MCL and CLL who relapsed while receiving ibrutinib.10-13 Similarly, activating mutations in phospholipase Cγ2, an immediate downstream target of BTK, were identified in patients receiving ibrutinib.12 Mutations that lead to activation of nonclassical NF-κB signaling were also reported in MCL and found to be associated with intrinsic resistance to ibrutinib.14 These mutations or other mechanisms that activate nonclassical NF-κB signaling may underlie a significant fraction of intrinsic ibrutinib failures. Thus, an optimal agent for ibrutinib-refractory MCL would overcome both BTK-resistance mutations and alternative NF-κB activation.
Heat shock protein 90 (HSP90) is a molecular chaperone that participates within multifactor complexes to stabilize client proteins and prevent their ubiquitination and proteasomal degradation.15 HSP90 is exploited by tumor cells to buffer malignancy-associated cellular stress16 and facilitate the maturation, activation, and stabilization of many oncoproteins.17 Inhibition of the adenosine triphosphate–dependent function of HSP90 with small-molecule inhibitors results in the dissociation of client proteins from the chaperone complex, with subsequent proteasomal degradation.16 We and others previously showed that targeting of JAK2 with newer-generation HSP90 inhibitors like AUY922 and PU-H71 is highly effective in both lymphoid and myeloid malignancies addicted to JAK2 signaling and can overcome resistance to enzymatic JAK2 inhibitors.18,19 This suggested that HSP90 inhibitors may overcome resistance through BTK C481S.
Although clinical activity of HSP90 inhibitors has been largely disappointing, there was a complete response to AUY922 in a patient with germinal center subtype diffuse large B-cell lymphoma following 2 cycles of therapy that has persisted for 24 months. In this trial, no patients with MCL were treated.20 We have initiated a Cancer Therapy Evaluation Program (CTEP)-sponsored phase 2 clinical trial of AT13387 (NCT02572453), a structurally similar HSP90 inhibitor to AUY922, in relapsed/refractory MCL resistant or intolerant to BTK inhibition, as well as BCL6-expressing large-cell lymphoma and anaplastic lymphoma kinase–positive anaplastic large-cell lymphoma. A central goal of this trial is to define the mechanisms that mediate both de novo and acquired resistance to HSP90 inhibitors, and thereby provide insights into the disparity between preclinical and clinical activity from this class of agents.
The specific clients that mediate response to HSP90 inhibition in MCL have not been defined. A recent HSP90 interactome performed in 293T cells quantified almost 400 client proteins and suggested that more than half of the human kinome can in principle interact with HSP90 and its cochaperone CDC37 at varying affinities.21 Significant differences exist across cell types in the complement of HSP90 clients. Thus, to more comprehensively define targets of HSP90 inhibition in MCL, we performed an agnostic proteomic analysis across MCL cell lines and xenografts.
Methods
MCL cell lines were purchased from ATCC and authenticated by short tandem repeat profiling. Primary MCL cells were cultured in RPMI 1640 (Pan Biotech) with 10% fetal bovine serum (FBS; Biochrom). Mouse splenic B cells were isolated by magnetic cell separation using α-CD43 (Ly-48) microbeads and LS columns to deplete CD43+ non-B cells from single-cell suspensions according to the manufacturer’s specifications (Miltenyi Biotec). Splenocytes from LMP1-CD40//CD19-cre mice were cultured in RPMI 1640 (Pan Biotech), supplemented with 2 mmol/L l-alanyl-glutamine (Gibco), 1 mmol/L sodium pyruvate (Gibco), 1× minimal essential medium nonessential amino acids (Gibco), 60 µM β-mercaptoethanol (Sigma-Aldrich), and 10% heat-inactivated FBS (Biochrom).
In vitro inhibitor assays
Viable cells were plated in white opaque 384-well plates (50 μL per well; Corning) using an EL406 Combination Washer Dispenser (BioTek) at a density of 0.05 × 106 to 0.1 × 106 cells per mL. Inhibitors or vehicle (dimethyl sulfoxide [DMSO]) were added using a JANUS Automated Workstation (PerkinElmer). For growth curve analysis, CellTiter-Glo 25 μL Luminescent Cell Viability reagent (Promega) was added at indicated time points to each well and read by the 2104 EnVision Multilabel Reader (PerkinElmer) as described.18 Each data point represents quadruplicates and experiments were repeated at least twice.
Flow cytometry
Apoptosis was determined using an fluorescein isothiocyanate Annexin V Apoptosis Detection Kit (BD Pharmingen) according to the manufacturer’s instructions in 500-μL aliquots of 106 cells per 4 mL treated as indicated. For cell-cycle analysis, separate 500-μL aliquots were stained with Hoechst 33342 dye for 1 hour at 37°C in the dark. Alternatively, 1.5 × 106 cells were washed and resuspended in ice-cold phosphate-buffered saline (PBS), added dropwise to 70% ethanol and frozen at −20°C for at least 12 hours. Fixed cells were then washed with ice-cold PBS, resuspended in 500 µL of propidium iodide (PI)/Triton X-100 staining solution (0.2 mg/mL RNAse A, 0.02 mg/mL PI, 0.1% Triton-X in PBS) and incubated for 20 minutes at 37°C. Flow cytometry was performed on a LSR Fortessa instrument (BD Biosciences) and analyzed with FlowJo software (Tristar). Flow cytometry for splenocytes from PDX mice used CD45-allophycocyanin from eBioscience, CD19-V450 from BD, and CD5-peridinin chlorophyll/Cy5.5 from Biolegend.
Multiplexed proteomics
Samples for multiplexed quantitative mass spectrometry (MS) analysis were processed and analyzed through the Thermo Fisher Scientific Center for Multiplexed Proteomics at Harvard Medical School.22 Sample processing steps included cell lysis, tandem protein digestion using LysC and trypsin, peptide labeling with Tandem Mass Tag 10-plex reagents and peptide fractionation. Multiplexed quantitative mass spectrometry data were collected on an Orbitrap Fusion mass spectrometer operating in MS3 mode using synchronous precursor selection for the MS2 to MS3 fragmentation.23 MS/MS data were searched against a Uniprot human database (February 2014) with both the forward and reverse sequences using the SEQUEST algorithm. Further data-processing steps included controlling peptide and protein level false discovery rates (FDRs), assembling proteins from peptides, and protein quantification from peptides.
Wild-type BTK and BTK C481S cloning and transfection
BTK wild type (wt) and BTK C481S were cloned into in pMSCV-ires-GFP and pMSCV-ires-tdTomato, respectively, using the Gateway vector conversion system as previously described.24 For retroviral production, we cotransfected BTK wt or BTK C481S and the retroviral packaging construct pVSVG (Addgene) at a 1:1 ratio into GP293 cells (Clontech) using Lipofectamine 2000 (Invitrogen). After 48 hours, we harvested the supernatant, passed it through a 0.45-μm filter (Millex; Millipore), and transduced 30 × 106 Jeko-1 or Mino-1 cells. After 1 day, we washed the cells and resuspended them in fresh media. Green fluorescent protein (GFP) and td-Tomato-positive cells were sorted twice on a FACSAria II sorter (BD Biosciences).
Transient expression of BTK in 293T
BTK (from PlasmID, Harvard Repository) was flanked with attB sites for Gateway-Cloning (Invitrogen) and tagged with 3xFlag as previously described.18 The C481S mutant was introduced by site-directed mutagenesis (QuikChange II XL; Stratagene). 293T cells were plated in a 10-cm dish (Corning) and transfected with 25 µg of plasmid DNA and 62.5 µL of Lipofectamine 2000 (Invitrogen). The next day, the cells were replated into 6-well plates, and after another 24 hours, incubated with inhibitors or vehicle (DMSO) at indicated doses and times.
Statistics
For proteomic data analysis, MS2 spectra were searched using the SEQUEST algorithm against a Uniprot composite database derived from the human proteome containing its reversed complement and known contaminants. Peptide spectral matches were filtered to a 1% FDR using the target-decoy strategy combined with linear discriminant analysis. Proteins from the 3 cell lines were filtered to a <0.01 FDR and were quantified from peptides with a summed signal to noise threshold of ≥200 and MS2 isolation specificity of 0.5. Normalized replicate samples in each cell line were averaged and quantified using ratios of treatment to control with log2 transformations to calculate fold change. Pearson product-moment correlation coefficients were used to assess the linear relationship between pairs of samples. Differential expression analyses were performed using the R package limma v3.26 within single and across multiple cell lines. Model statistics were computed based on ordinary and moderated standard errors using an empirical Bayes method to shrink sample variances due to few samples. Proteins that met fold change and FDR cutoffs in both parental, BTK wt and C481S transduced and PDX samples were selected, and proteins appearing in at least 4 of 5 comparisons were selected for further analysis. Proteins appearing in <4 were also summarized. The cutoff value of log2 fold change for upregulated or downregulated proteins was 0.5. Statistical analysis of quantified proteins was performed using R (version 3.2) and visualized in the form of distribution, volcano, and violin blots. Heatmaps were generated using GENE-E (The Broad Institute of MIT and Harvard, version 3.0.204). Classification and pathway membership were analyzed using Ingenuity Pathway analysis (Qiagen). Kinome dendrogram illustration was an adapted and reproduced courtesy of Cell Signaling Technology, Inc using Kinome render (http://bcb.med.usherbrooke.ca/kinomerender.php). Dose-response curves, 50% inhibitory concentration (IC50), and plots were generated with GraphPad Prism software. Continuous variables were compared with 2-sided Student t tests.
Study approval
The acquisition and use of primary specimens was approved by Dana-Farber Cancer Institute (DFCI) Institutional Review Board #01-206. All animal studies were performed under DFCI Animal Care Use Committee protocol #13-351.
Additional methods are available in supplemental Methods (see supplemental Data, available on the Blood Web site).
Results
AUY922 overcomes nonclassical NF-κB signaling and BTK C481S
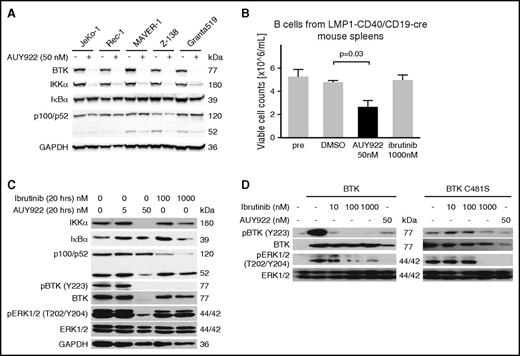
All cases of MCL are believed to be dependent on either classical or nonclassical NF-κB signaling, with the former activated through the BCR and the latter conferring intrinsic resistance to BTK inhibition.14 Treatment of MCL cell lines with AUY922 (50 nM) caused the near complete depletion of both BTK and IκB kinase α (IKKα) proteins in all 5 lines (Figure 1A). Importantly, this concentration of AUY922 50 nM is far below the serum concentrations achievable in humans.25 Among the 5 MCL lines, Z-138 harbors a nonsense mutation in TRAF2 and MAVER-1 has a deletion of TRAF3. Both of these mutations result in activation of the nonclassical NF-κB pathway.14 In both lines, as well as Granta519 cells that lack a known activating mutation of nonclassical NF-κB, depletion of IKKα was associated with reduced cleavage of p100/NFKB2 to the active form p52 (Figure 1A). Thus, HSP90 inhibition can suppress downstream signaling through nonclassical NF-κB in MCL.
HSP90 inhibition targets BTK and IKKα to overcome BTK resistance. (A) Immunoblotting of MCL lines for the indicated targets after 48-hour exposure to AUY922 50 nM or vehicle. (B) Viable cell counts of splenic B cells from LMP1-CD40//CD19-Cre mice prior to exposure (pre) or upon exposure to the indicated agents for 20 hours. P value based on 2-sided unpaired Student t test. Error bars indicate standard deviation (SD). (C) Immunoblotting of the cells described in panel B for the indicated targets. (D) Immunoblotting of 293T cells expressing wt BTK or BTK C481S and treated for 20 hours with the indicated agents. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
HSP90 inhibition targets BTK and IKKα to overcome BTK resistance. (A) Immunoblotting of MCL lines for the indicated targets after 48-hour exposure to AUY922 50 nM or vehicle. (B) Viable cell counts of splenic B cells from LMP1-CD40//CD19-Cre mice prior to exposure (pre) or upon exposure to the indicated agents for 20 hours. P value based on 2-sided unpaired Student t test. Error bars indicate standard deviation (SD). (C) Immunoblotting of the cells described in panel B for the indicated targets. (D) Immunoblotting of 293T cells expressing wt BTK or BTK C481S and treated for 20 hours with the indicated agents. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
To determine whether HSP90 inhibition has an effect on nonclassical NF-κB pathway function, we used transgenic murine B cells with constitutive activation of the nonclassical NF-κB pathway downstream of CD40.26 Treatment of splenic B cells from LMP1-CD40/CD19-cre mice with AUY922 (50 nM) reduced cell viability (Figure 1B). As expected, ibrutinib (1000 nM) had no effect (Figure 1B). As in human MCL cells, AUY922 (50 nM) resulted in the complete degradation of IKKα and markedly reduced p100 cleavage whereas ibrutinib appeared to increase p100 cleavage (Figure 1C). Treatment with AUY922 also markedly reduced BTK protein and phosphorylation of its downstream target extracellular signal-regulated kinase (ERK1/2) in these cells whereas the effect of ibrutinib was limited to reduced phosphorylation of BTK (Figure 1C).
Mutations in JAK2, epidermal growth factor receptor (EGFR), and BCR-ABL that confer resistance to type I and type II inhibitors do not reduce HSP90 inhibitor sensitivity.18,27,28 In fact, these resistance mutations may contribute to protein instability and thus increase dependence on HSP90 chaperone activity. To test the effects of BTK C481S on HSP90 dependence, we used 293T cells, which do not express endogenous BTK. Ectopic expression of either wt BTK or BTK C481S resulted in constitutive phosphorylation of BTK and ERK1/2 (Figure 1D). As expected, ibrutinib (10 nM) almost completely abrogated BTK phosphorylation in cells expressing wt BTK but ibrutinib (100 nM) had no effect on the phosphorylation of BTK C481S (Figure 1D). In contrast, treatment with AUY922 (50 nM) resulted in downregulation of BTK and complete loss of phosphorylated ERK1/2 (pERK1/2) in cells expressing either wt BTK or BTK C481S (Figure 1D). Thus, potent inhibition of HSP90 may overcome both intrinsic resistance to ibrutinib through the nonclassical NF-κB pathway and acquired resistance from BTK C481S.
Agnostic identification of HSP90 clients in MCL
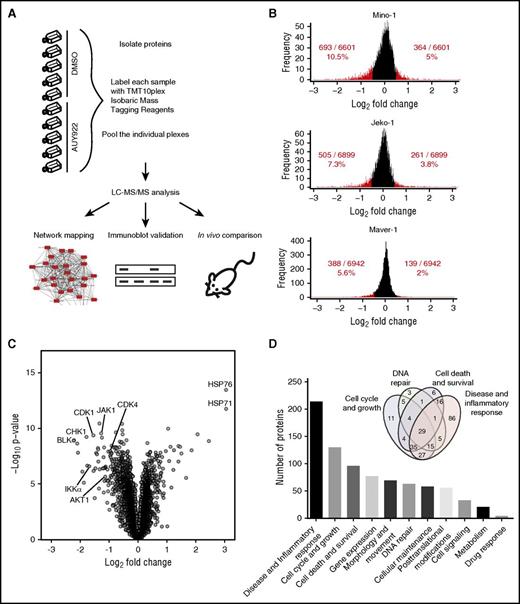
Recent advances in multispecimen labeling and high-sensitivity proteomics have made it possible to compare multiple admixed samples for protein abundance across the proteome. We used high-resolution Orbitrap mass spectrometers and isobaric tandem mass tags to quantify whole proteomes from lysates of Mino, JeKo-1, and MAVER-1 cells treated with either DMSO or AUY922 (Figure 2A). We chose these lines based on their differential genetics and sensitivity to ibrutinib (Table 1). Differences in the abundance of nearly 7000 proteins were quantified between treatment with AUY922 or DMSO, with a similar distribution of protein level fold change for each cell line (Figure 2B; supplemental Table 1). There was approximately a twofold difference in percent of downregulated compared with upregulated proteins using log2 fold change ≥0.5 as a cutoff (Figure 2B). Among the most highly downregulated were proteins specifically associated with MCL, including CDK4, IKKα, and AKT (Figure 2C). As expected,29 the most highly upregulated proteins included members of the HSP70 family, HSP76 and HSP71 (Figure 2C). Analysis of dysregulated proteins using Ingenuity software revealed that over 50% of the proteins with log2 fold change of ≥0.5 are involved in cell-cycle and growth, disease and inflammatory response, cell death and survival, and/or DNA repair (Figure 2D; supplemental Table 2). Ingenuity pathway analysis of dysregulated proteins identified NF-κB, BCR signaling, and DNA repair among the most enriched pathways (supplemental Table 3).
In-depth quantitative proteomics of MCL cell lines treated with HSP90 inhibition. (A) Experimental strategy. Lysates from MCL cell lines Mino, JeKo-1, and MAVER-1 (parental and/or transduced to express either wt BTK or BTK C481S) treated with either DMSO or AUY922 50 nM for 24 hours were used for protein isolation and digestion. Peptides were labeled with Tandem Mass Tag 10-plex (TMT10plex) reagents and peptide fractionation. Multiplexed quantitative MS data were collected and analyzed. (B) Comparison between protein levels in parental and transduced Mino, JeKo-1, and Maver-1 cell lines for DMSO- vs AUY922-treated cells. Proteins downregulated and upregulated by log2 fold ≥0.5 are represented in red. (C) Volcano plot demonstrating the distribution of protein fold changes between parental and transduced Mino, JeKo-1, and Maver-1 cell lines for AUY922- vs DMSO-treated cells, highlighting the most highly upregulated and downregulated proteins following HSP90 inhibition with AUY922. (D) Pathway analysis using Ingenuity pathway analysis (Qiagen) identifying the cellular and molecular pathways most highly affected by protein level changes following HSP90 inhibition with AUY922 in MCL lines. LC, liquid chromatography.
In-depth quantitative proteomics of MCL cell lines treated with HSP90 inhibition. (A) Experimental strategy. Lysates from MCL cell lines Mino, JeKo-1, and MAVER-1 (parental and/or transduced to express either wt BTK or BTK C481S) treated with either DMSO or AUY922 50 nM for 24 hours were used for protein isolation and digestion. Peptides were labeled with Tandem Mass Tag 10-plex (TMT10plex) reagents and peptide fractionation. Multiplexed quantitative MS data were collected and analyzed. (B) Comparison between protein levels in parental and transduced Mino, JeKo-1, and Maver-1 cell lines for DMSO- vs AUY922-treated cells. Proteins downregulated and upregulated by log2 fold ≥0.5 are represented in red. (C) Volcano plot demonstrating the distribution of protein fold changes between parental and transduced Mino, JeKo-1, and Maver-1 cell lines for AUY922- vs DMSO-treated cells, highlighting the most highly upregulated and downregulated proteins following HSP90 inhibition with AUY922. (D) Pathway analysis using Ingenuity pathway analysis (Qiagen) identifying the cellular and molecular pathways most highly affected by protein level changes following HSP90 inhibition with AUY922 in MCL lines. LC, liquid chromatography.
Relative IC50 (µM) for MCL lines treated with the indicated agents for 48 hours
| . | Mino . | JeKo-1 . | Z-138 . | Maver-1 . | Rec-1 . | Granta519 . |
|---|---|---|---|---|---|---|
| AUY922 | 0.012 | 0.0068 | 0.0039 | 0.0038 | 0.0075 | 0.006 |
| AT13387 | 0.13 | 0.075 | 0.065 | 0.33 | 0.13 | 0.13 |
| PU-H71 | 0.38 | 0.072 | 0.04 | 0.073 | 0.29 | 0.11 |
| HSP990 | 0.48 | 11 | 0.005 | 0.011 | 0.021 | 0.24 |
| 17-AAG | 0.51 | 0.15 | 0.029 | 0.14 | 10 | 0.67 |
| Ibrutinib | 6.8 | 2.7 | 12 | 6.6 | 6.5 | 22 |
| ABT737 | ND | 1.0 | 0.0046 | 0.017 | 0.26 | 0.12 |
| Bortezomib | ND | 0.002 | 0.002 | 0.0025 | 0.0031 | 0.0013 |
| Bendamustine | ND | 160 | 36 | 150 | 490 | 3000 |
| Idelalisib | ND | 6.9 | 7.7 | 27 | 67 | 19 |
| Cytarabine | ND | 0.03 | 0.016 | 0.022 | 0.07 | 0.14 |
| Doxorubicin | ND | 0.098 | 0.0069 | 0.057 | 0.088 | 0.015 |
| Ifosfamide | ND | 1200 | 400 | 540 | 1200 | 690 |
| Hydrocortisone | ND | 550 | 320 | 860 |
| . | Mino . | JeKo-1 . | Z-138 . | Maver-1 . | Rec-1 . | Granta519 . |
|---|---|---|---|---|---|---|
| AUY922 | 0.012 | 0.0068 | 0.0039 | 0.0038 | 0.0075 | 0.006 |
| AT13387 | 0.13 | 0.075 | 0.065 | 0.33 | 0.13 | 0.13 |
| PU-H71 | 0.38 | 0.072 | 0.04 | 0.073 | 0.29 | 0.11 |
| HSP990 | 0.48 | 11 | 0.005 | 0.011 | 0.021 | 0.24 |
| 17-AAG | 0.51 | 0.15 | 0.029 | 0.14 | 10 | 0.67 |
| Ibrutinib | 6.8 | 2.7 | 12 | 6.6 | 6.5 | 22 |
| ABT737 | ND | 1.0 | 0.0046 | 0.017 | 0.26 | 0.12 |
| Bortezomib | ND | 0.002 | 0.002 | 0.0025 | 0.0031 | 0.0013 |
| Bendamustine | ND | 160 | 36 | 150 | 490 | 3000 |
| Idelalisib | ND | 6.9 | 7.7 | 27 | 67 | 19 |
| Cytarabine | ND | 0.03 | 0.016 | 0.022 | 0.07 | 0.14 |
| Doxorubicin | ND | 0.098 | 0.0069 | 0.057 | 0.088 | 0.015 |
| Ifosfamide | ND | 1200 | 400 | 540 | 1200 | 690 |
| Hydrocortisone | ND | 550 | 320 | 860 |
Data represent the mean of 2 to 3 independent experiments performed in triplicate.
ND, not determined.
Next, we asked whether expression of BTK C481S alters the MCL proteome. We expressed either wt BTK or BTK C481S in Mino and Jeko-1 cells and performed agnostic proteome analysis in the presence or absence of AUY922. Expression of neither wt BTK nor BTK C481S had significant effects on the overall proteome (supplemental Figure 1). Compared with parental cells, cells expressing BTK C481S had only 40 proteins that were altered by a log2 fold change of ≥0.5 in the presence of AUY922 in both lines; none were known drivers of MCL pathogenesis (supplemental Table 4).
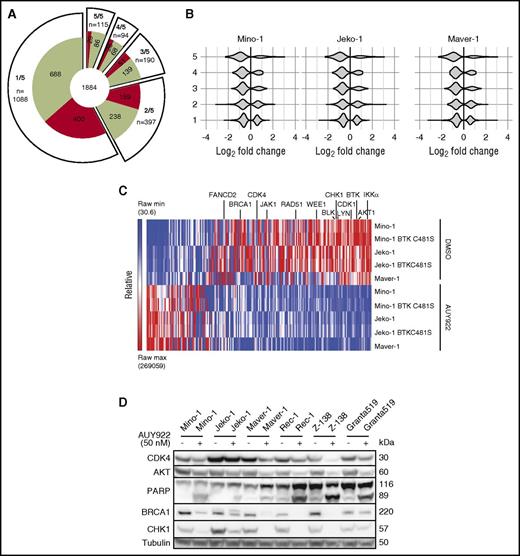
To identify consistently affected proteins by AUY922 across conditions, we compared 5 experiments (DMSO vs AUY922 for Mino, Jeko-1, Maver1, Mino expressing BTK C481S and Jeko expressing BTK C481S) and quantified the number of proteins that were altered by a log2 fold change of ≥0.5 with AUY922 treatment in 1, 2, 3, 4, or all 5 comparisons (Figure 3A). In total, 1884 proteins were altered by a log2 fold change of ≥0.5 in at least 1 comparison with AUY922 treatment (65% downregulated), with 209 proteins observed in at least 4 of 5 comparisons (Figure 3A; supplemental Table 4). Of the 209, 74% were downregulated and 26% were upregulated (P < .001). The distribution of protein changes in each cell line shifted toward a more downregulated profile as we considered proteins whose levels were significantly altered in multiple different cell lines (Figure 3B).
Consistent targets of HSP90 inhibition across MCL lines. (A) Distribution of proteins that were upregulated (red) or downregulated (green) by log2 fold ≥0.5 following treatment with AUY922 in 1 of 5, 2 of 5, 3 of 5, 4 of 5, and 5 of 5 cell lines. (B) Violin plots of protein fold changes for proteins that were altered by log2 fold ≥0.5 in 1 to 5 cell lines among Mino, JeKo-1, and Maver-1 cell lines following treatment with AUY922 vs DMSO. (C) Heatmap demonstrating protein levels of the most highly regulated proteins by HSP90 inhibition. Proteins were included if they were up or downregulated by log2 fold ≥0.5 in at least 4 of 5 cell lines, demonstrating an abundance of downregulated targets including proteins involved in BCR and JAK/STAT signaling, the nonclassical NF-κB pathway, and DNA repair and cell-cycle control. (D) Immunoblotting of MCL lines for the indicated targets after 48-hour exposure to AUY922 50 nM or vehicle.
Consistent targets of HSP90 inhibition across MCL lines. (A) Distribution of proteins that were upregulated (red) or downregulated (green) by log2 fold ≥0.5 following treatment with AUY922 in 1 of 5, 2 of 5, 3 of 5, 4 of 5, and 5 of 5 cell lines. (B) Violin plots of protein fold changes for proteins that were altered by log2 fold ≥0.5 in 1 to 5 cell lines among Mino, JeKo-1, and Maver-1 cell lines following treatment with AUY922 vs DMSO. (C) Heatmap demonstrating protein levels of the most highly regulated proteins by HSP90 inhibition. Proteins were included if they were up or downregulated by log2 fold ≥0.5 in at least 4 of 5 cell lines, demonstrating an abundance of downregulated targets including proteins involved in BCR and JAK/STAT signaling, the nonclassical NF-κB pathway, and DNA repair and cell-cycle control. (D) Immunoblotting of MCL lines for the indicated targets after 48-hour exposure to AUY922 50 nM or vehicle.
Among the 209 proteins whose levels were significantly altered by AUY922 in 4 or 5 (n = 94) or all 5 of the 5 (n = 115) comparisons, only 26 were kinases (supplemental Figure 2). Of the 26, 25 were downregulated and included BCR-signaling factors LYN, BTK, and BLK, IKKα, JAK1, and AKT1 (Figure 3C). Thus, consistent targets of HSP90 inhibition on the MCL kinome include many that are known dependences in this disease. Also among the 209 proteins were factors essential for homology-directed repair, including BRCA1, RAD51, CHK1, WEE1, and FANCD2 (Figure 3C). Many of these proteins have previously been identified as HSP90-dependent but not in a robust fashion across multiple cell lines from the same disease.30,31 We confirmed the downregulation of multiple factors by immunoblotting across multiple lines (Figure 3D). Importantly, serum starvation of MAVER-1 cells for 48 hours to induce cell-cycle arrest had little or no effect on levels of BRCA1, IKKα, CHK1, and CDK4 (supplemental Figure 3), suggesting that cell-cycle alterations are not the driver of protein alterations from HSP90 inhibition in this context.
Activity of HSP90 inhibitors in MCL
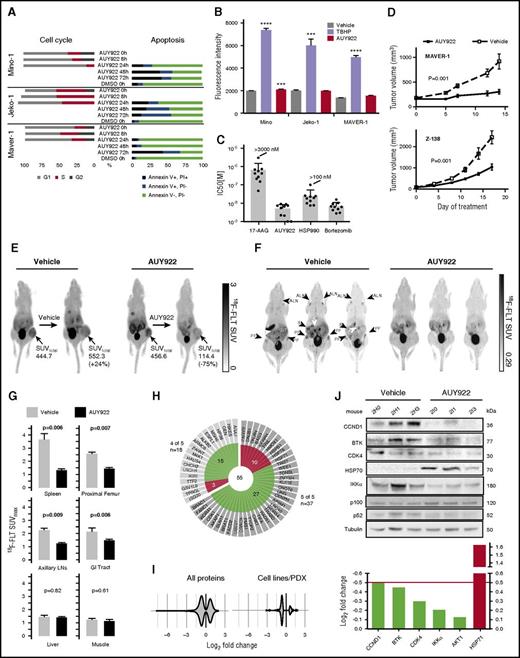
We assayed 5 structurally diverse HSP90 inhibitors (supplemental Figure 4), including the geldanamycin-derivative 17-AAG and 4 newer-generation drugs (AUY922, PU-H71, HSP990, and AT13387), across a panel of MCL lines. The newer-generation HSP90 inhibitors had 10- to 100-fold higher potency compared with 17-AAG in standard proliferation assays (Table 1). These HSP90 inhibitors were also 10- to 10 000-fold more potent than other agents used in the treatment of B-cell lymphomas, including cytotoxic chemotherapies, the PI3K inhibitor idelalisib, and ibrutinib (Table 1). AUY922 (50 nM) induced G0/G1 cell-cycle arrest within 8 to 24 hours and apoptosis between 24 and 72 hours in Mino, Jeko-1 and MAVER-1 (Figure 4A).
Newer-generation HSP90 inhibitors potently inhibit MCL in vitro and in vivo. (A) Cell-cycle analysis by flow cytometry upon treatment with AUY922 50 nM after 0, 8, and 24 hours, and Annexin V (AV) and PI staining of MCL lines treated as indicated. Data are representative of 2 to 4 independent experiments. (B) ROS as measured by DCFDA fluorescence intensity at 3 hours for Mino, Jeko-1, and MAVER-1 cell lines cultured with DCFDA along with vehicle, TBHP (positive control), or AUY922. Error bars indicate SD. ***P < .001, ****P < .0001. (C) IC50 values for 10 primary MCL samples. (D) Mice were inoculated subcutaneously with either MAVER-1 or Z-138 (n = 40 per line) and treated with AUY922 or vehicle once a volume of 100 to 200mm3 was reached. Error bars indicate standard error of the mean (SEM). P = .001 for both MAVER-1 and Z-138 compared with vehicle by 2-way ANOVA. (E) 18F-FLT PET imaging of mice with Z-138 xenografts treated for 5 days. SUVtotal indicates the standard uptake value integrated over the entire tumor volume. (F) Images from 18F-FLT PET in NSG mice xenografted with primary patient MCL and treated with either vehicle or AUY922 for 5 days. (G) Average SUVmax (n = 3 per arm) for the indicated organs. Proximal femur and lymph nodes each represent averages of right and left within the same animal. P values based on 2-sided Student t test. Error bars indicate SD. (H) Distribution of proteins that were upregulated (red) or downregulated (green) by log2 fold ≥0.5 following treatment with AUY922 in mouse PDX models and 4 of 5 or 5 of 5 cell lines. (I) Violin plots of protein fold changes for proteins that were altered by log2 fold ≥0.5 in the mouse PDX model alone and in both the mouse PDX model and 1 to 5 cell lines following treatment with AUY922 vs DMSO. (J) Immunoblotting of lysates from spleens of mice xenografted with primary MCL and treated for 5 days, and log fold change of each of protein represented in the immunoblot from the proteomic experiment. ALN, axillary lymph node; B, bladder; GIT, gastrointestinal tract; K, kidney; PF, proximal femur; S, spleen.
Newer-generation HSP90 inhibitors potently inhibit MCL in vitro and in vivo. (A) Cell-cycle analysis by flow cytometry upon treatment with AUY922 50 nM after 0, 8, and 24 hours, and Annexin V (AV) and PI staining of MCL lines treated as indicated. Data are representative of 2 to 4 independent experiments. (B) ROS as measured by DCFDA fluorescence intensity at 3 hours for Mino, Jeko-1, and MAVER-1 cell lines cultured with DCFDA along with vehicle, TBHP (positive control), or AUY922. Error bars indicate SD. ***P < .001, ****P < .0001. (C) IC50 values for 10 primary MCL samples. (D) Mice were inoculated subcutaneously with either MAVER-1 or Z-138 (n = 40 per line) and treated with AUY922 or vehicle once a volume of 100 to 200mm3 was reached. Error bars indicate standard error of the mean (SEM). P = .001 for both MAVER-1 and Z-138 compared with vehicle by 2-way ANOVA. (E) 18F-FLT PET imaging of mice with Z-138 xenografts treated for 5 days. SUVtotal indicates the standard uptake value integrated over the entire tumor volume. (F) Images from 18F-FLT PET in NSG mice xenografted with primary patient MCL and treated with either vehicle or AUY922 for 5 days. (G) Average SUVmax (n = 3 per arm) for the indicated organs. Proximal femur and lymph nodes each represent averages of right and left within the same animal. P values based on 2-sided Student t test. Error bars indicate SD. (H) Distribution of proteins that were upregulated (red) or downregulated (green) by log2 fold ≥0.5 following treatment with AUY922 in mouse PDX models and 4 of 5 or 5 of 5 cell lines. (I) Violin plots of protein fold changes for proteins that were altered by log2 fold ≥0.5 in the mouse PDX model alone and in both the mouse PDX model and 1 to 5 cell lines following treatment with AUY922 vs DMSO. (J) Immunoblotting of lysates from spleens of mice xenografted with primary MCL and treated for 5 days, and log fold change of each of protein represented in the immunoblot from the proteomic experiment. ALN, axillary lymph node; B, bladder; GIT, gastrointestinal tract; K, kidney; PF, proximal femur; S, spleen.
To assess the contribution of reactive oxygen stress (ROS) from HSP90 inhibition, we cultured Mino, Jeko-1, and MAVER-1 cells with the fluorescent ROS reporter 2′7′-dichlorofluorescin diacetate (DCFDA) in the presence of either tert-butyl hydroperoxide (TBHP) (positive control), vehicle (negative control), or AUY922. There was approximately a threefold increase in ROS in the presence of TBHP in each line (Figure 4B). In contrast, treatment with AUY922 led to a minimal absolute increase in ROS in each line (Figure 4B). Culturing of Jeko-1 cells in AUY922 (10 nM) for 72 hours had no effect on ROS (data not shown).
To confirm the activity of HSP90 inhibition in primary human MCL, we assayed a panel of 10 patient samples from pleural effusion or peripheral blood specimens (see supplemental Methods). All 10 samples were uniformly sensitive to AUY922 (IC50, 1.0-9.0 nM) and bortezomib (IC50, 2.8-15.9 nM) (Figure 4C). Importantly, peripheral blood mononuclear cells from 2 healthy individuals were not sensitive to AUY922 (IC50 >100 nm) (supplemental Table 5).
To assay the activity of AUY922 against MCL in vivo, we xenografted MAVER-1 (p53 mutant, TRAF3-deleted) and Z-138 cells (p53 wt, TRAF2-mutant) into SCID-beige mice.14 AUY922 50 mg/kg by tail vein injection thrice weekly significantly reduced the growth of both MAVER-1 and Z-138 xenografts compared with vehicle (P = .001 each by 2-way analysis of variance [ANOVA]; Figure 4D). We performed 18F-fluorothymidine (18F-FLT) positron emission tomography (PET) scanning on mice xenografted with Z-138 after 5 days of treatment with AUY922 or vehicle. 18F-FLT PET can sensitively detect early responses to targeted therapy in murine lymphoma xenografts32 and correlates with inhibition of CDK4 activity in humans treated with the CDK4 inhibitor PD0332991.33 Treatment with AUY922 reduced tumor PET activity by 75% compared with the same mouse prior to AUY922 treatment (Figure 4E). In contrast, tumor PET activity increased within the same mouse by 24% over 5 days of vehicle treatment (Figure 4E). Thus, HSP90 inhibition has activity against MCL in vivo, including lines with intrinsic ibrutinib resistance through constitutive activation of nonclassical NF-κB.34
To further analyze the effect of HSP90 inhibition on primary MCL in vivo, we xenografted a peripheral blood sample into Nod.Scid.IL2Rγ−/− (NSG) mice from a patient who had failed multiple lines of chemotherapy as well as ibrutinib (see supplemental Methods). Within 28 days, fatal CD5+CD19+CD20+ κ-restricted MCL developed in the spleen, bone marrow, liver, and intestines (supplemental Figure 5). Xenografted MCL cells from the spleen were secondarily transplanted into NSG mice (n = 3 per arm), which were randomized to receive either AUY922 50 mg/kg or vehicle by tail-vein injection thrice weekly. 18F-FLT PET after 5 days of treatment with AUY922 showed significantly reduced PET activity in spleen, bone marrow, lymph nodes, and intestines compared with vehicle-treated mice (Figure 4F-G). In contrast, PET activity in tissues uninvolved by MCL (eg, muscle) was unaffected by AUY922.
Finally, we performed agnostic proteomic analysis of spleens collected from mice engrafted with the MCL PDX and treated with AUY922 or vehicle for 5 days (supplemental Table 6). Spleens from mice treated with AUY922 for 5 days had ∼60% involvement with MCL compared with >95% involvement in vehicle-treated spleens (data not shown). Of the 209 proteins altered by log2 fold change ≥0.5 by AUY922 treatment in 4 of 5 cell-line conditions (Figure 3A). 55 were also altered by log2 fold ≥0.5 in the in vivo xenograft (Figure 4H). These 55 included many of the previously noted proteins involved in DNA repair and cell-cycle control: WEE1, RAD51, CHK1, CDK1, FANC1, and BRCA1. Again, the distribution of protein in the PDX shifted toward a more downregulated profile as we considered only proteins affected in both the cell lines and xenograft proteomics discovery (Figure 4I). Of note, BTK, CDK4, and IKKα did not score in the proteomics analysis but immunoblotting of involved spleens from mice treated for 5 days showed downregulation of all 3 in AUY922-treated MCL cells compared with vehicle-treated (Figure 4J). Loss of IKKα was associated with reduced p100 cleavage to p52 (Figure 4J), indicating that AUY922 can inhibit nonclassical NF-κB signaling in primary human MCLs in vivo.
Discussion
The efficacy of BTK inhibition in MCL has been the most exciting development in the treatment of this disease in recent years. However, many patients exhibit de novo resistance to ibrutinib and median survival following progression on ibrutinib is <1 year.9 The BTK C481S mutation appears to be less common in patients with MCL than in those with CLL, indicating that alternate mechanisms remain to be elucidated. Using a robust, agnostic proteomic approach, we demonstrated across multiple MCL lines in vitro, and in PDX models of this disease in vivo, that inhibition of HSP90 results in the downregulation of a restricted set of kinases and DNA repair factors involved in survival and proliferation in MCL. MCL is defined by overexpression of cyclin D1 and activation of CDK4/6, BCR signaling, and NF-κB signaling, and components of each of these pathways were among the most highly downregulated across multiple cell lines, as well as in vivo.
Although our analysis discovered a majority of previously reported HSP90 clients, there was a notable difference in the effect of HSP90 inhibition on the kinome of MCL cell lines compared with what has been previously reported in an HSP90 interactome.21 Whereas this analysis revealed protein kinases to be among the most strongly associated proteins that were bound by HSP90, accounting for over 50% of HSP90 clients discovered by this methodology, our analysis found only 26 kinases to be significantly affected by inhibition of HSP90, accounting for 12% of significantly affected proteins. Although tyrosine kinases as well as cyclin-dependent kinases, MAPKs, and glycogen synthetase kinases were well represented in both, calcium-calmodulin–dependent kinases, AGC group kinases, and serine-threonine kinases were notably not affected in MCL lines. This suggests that relevant interactions are likely to differ across cell types and diseases, thereby influencing the efficacy of HSP90 in each context. It is also important to consider that secondary responses to HSP90 inhibition, such as the unfolded protein response, may also drive activity in preclinical models. Although we did not find compelling evidence for this in our pathway analysis (supplemental Table 3), in vivo systems are likely to be important for distinguishing direct target effects from downstream consequences.
There has been interest in HSP90 inhibition for the treatment of malignancies for almost 2 decades. Although these drugs have shown activity in some patients, they have been generally disappointing. Early phase studies of HSP90 inhibitors in solid tumors showed that, despite a long intratumoral half-life, the effect of HSP90 inhibitors on the downregulation of client proteins is short-lived.35 This may be the result of compensatory upregulation of other heat shock proteins such as HSP70 and heat shock factor-1 (HSF-1).36,37 Additionally, alterations in systemic and intratumoral drug metabolism may play a role, as has been suggested by the correlation between response and polymorphisms in uridine 5′-diphospho-glucuronosyltransferace in colorectal and bladder cancer cell lines.38,39 Finally, resistance to HSP90 inhibition has been linked to defects in apoptosis. In some contexts, sensitivity to HSP90 inhibitors is reliant on the proapoptotic protein BAX,40,41 as well as on drug-induced downregulation of the antiapoptotic protein MCL1.41 Loss of dependency on MCL1 for cell survival has been shown to result in resistance to HSP90 inhibition in vitro, but may be overcome by combination therapy with agents targeting BCL2, BCLxL, and BCL-w.41
Whether 1 or more of these mechanisms mediate resistance in MCL remains unclear, as does the most effective combination strategy to overcome in vivo resistance. For example, bromodomain and CDK7 inhibitors may be effective against the transcriptional upregulation of HSF-1 and its targets. Similarly, the downregulation of proteins involved in homology-directed repair by AUY922 in our proteomic studies and similarly reported by others30,42,43 suggests a possible synergy between inhibitors of poly (ADP-ribose) polymerase 1 (PARP1) and HSP90. In fact, this combination will be evaluated in an upcoming phase 1 clinical trial of talazoparib and AT13387 in patients with metastatic advanced recurrent ovarian, fallopian tube, primary peritoneal, or triple-negative breast cancer (NCT02627430).
Rather than proceed to combinations based on preclinical data, we believe the most important next step is to more fully understand the discrepancy between the preclinical and clinical efficacy of single-agent HSP90 inhibitors. Based on our demonstrated efficacy of HSP90 inhibition in ibrutinib-sensitive and ibrutinib-resistant MCL cell lines, primary samples and a PDX, we have initiated a phase 2 clinical trial of AT13387 in relapsed/refractory MCLs resistant or intolerant to BTK inhibition (NCT02572453). Extensive correlative studies are required for enrolling patients, including pretreatment, on-treatment, and postrelapse biopsies. The data set we outline here serves as the predicted set of on-target biomarkers. Mechanisms of resistance will be prospectively determined both in patients with upfront failure and in those with progression after response. We believe this rational approach to elucidating HSP90 inhibitor activity will help define the most appropriate combinations that can maximize their clinical benefit for patients with MCL and possibly other cancers.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Thomas Radimerski, Frank Stegmeier, Ryan Kunz, Samuel Newmark, and Quang-de Nguyen for assistance and thoughtful comments.
This work was supported by the Max-Eder Program of the Deutsche Krebshilfe (#110659 [O.W.]), the German Research Foundation (SFB1243 TP-A11 [O.W.] and TP-A13 [U.Z.-S.]), the William Lawrence and Blanche Hughes Foundation (D.M.W.), and a Leukemia & Lymphoma Society Translational Research Program award (D.M.W.).
Authorship
Contribution: N.K., C.J., O.W., and D.M.W. contributed to the experiments and preparation of the manuscript; R.A.R. contributed to the statistical analysis and review of the manuscript; and J.V.L., S.T., S.H., D.v.B., L.B., A.L.C., A.C., A.S., T.T., S.Z., D.B., U.Z.-S., and S.J.R. contributed to the experiments and the review of the manuscript.
Conflict-of-interest disclosure: D.M.W. received research support and consulting fees from Novartis. The remaining authors declare no competing financial interests.
Correspondence: David M. Weinstock, Dana-Farber Cancer Institute, 450 Brookline Ave, Dana 510B, Boston, MA 02215; e-mail: dweinstock@partners.org; and Oliver Weigert, Ludwig-Maximilians University, Munich, Max-Lebsche Platz 30, D3/208, 81377 Munich, Germany; e-mail: oliver.weigert@med.uni-muenchen.de.
References
Author notes
C.J. and N.K. contributed equally.
O.W. and D.M.W. contributed equally.