Key Points
Expression of miR-139-5p is downregulated in BCR-ABL–mediated CML patients.
miR-139-5p regulates proliferation and differentiation activities by targeting Brg1 in early hematopoiesis.
Abstract
MicroRNAs (miRNAs) have emerged as important regulators of the immune system. However, despite this prominence, our understanding of the function of miRNAs in the early hematopoietic stages is incomplete. In this study, we found that miR-139-5p negatively regulated the proliferation of hematopoietic stem cells and progenitor cells and that downregulation of miR-139-5p expression was associated with hematopoietic malignancy, such as chronic myeloid leukemia (CML). Knockdown of miR-139-5p resulted in myeloid-biased differentiation with expansion of myeloid progenitor cells. In contrast, miR-139-5p expression inhibited the proliferation of hematopoietic progenitors and resulted in the remission of a CML-like disease that is induced by breakpoint cluster region–Abelson (BCR-ABL) transformation. We also found that Brg1 is a functional target of miR-139-5p and that Brg1 is involved in BCR-ABL–induced leukemogenesis. Thus, our results identify miR-139-5p as a key regulator of cellular proliferation during early hematopoiesis and suggest that it is a potent antileukemic molecule.
Introduction
Hematopoiesis is a hierarchical stepwise process of lineage commitment, proliferation, and differentiation to generate distinct blood cells in the immune system. Hematopoietic stem cells (HSCs) are capable of self-renewal and give rise to multilineage progenitor cells or single lineage-restricted precursor cells.1 The balance between the self-renewal capacity and differentiation of HSCs is crucial to maintain the pool of stem cells and to replenish mature blood cells. Leukemia can arise from leukemic stem cells (LSCs) that are analogous to normal HSCs and progenitor cells. Because LSCs are capable of self-renewal for long-term clonal maintenance and give rise to differentiated myeloid lineage cells, they appear to arise from genetic mutation and/or selective gene expression of normal HSCs or more committed progenitor cells.2 Thus, understanding the genetic programs that control self-renewal and lineage commitment in HSCs and progenitor cells would provide a detailed insight into hematopoietic differentiation and yield new perspectives for preventive and therapeutic strategies for hematopoietic malignancies.
MicroRNAs (miRNAs) regulate timing and gene expression levels during developmental and physiological processes, including proliferation, apoptosis, and differentiation.3 miRNAs, which are ∼22 nucleotides (nt) in length, regulate gene expression at the posttranscriptional level by degrading their target messenger RNAs (mRNAs) or inhibiting translation of mRNAs by binding to the complementary seed sequences within the 3′ untranslated region (UTR) of target mRNAs.4 Previous reports have shown that miRNAs are important players in the development and differentiation of the hematopoietic system. Deletion of factors involved in the generation and function of miRNAs, such as Argonaute2 and Dicer, can cause defects in immune cell development.5,6 In addition, many studies have identified specific miRNAs with important roles in early hematopoiesis. For example, miR-126 controls HSC maintenance, and increased expression of miR-29a promotes aberrant HSC proliferation.7-9 miR-125a/b expression is associated with HSC activity and leukemogenesis.10-12 Recently, miR-139 has been associated with HSC function and leukemia. Ectopic miR-139-5p expression inhibits protein translation in acute myeloid leukemia (AML), and miR-139-3p inhibits the proliferation of myeloid progenitors.13,14 However, the physiological function and the molecular mechanism of miR-139-5p in early hematopoiesis and other types of leukemia, such as chronic myeloid leukemia (CML), still remain largely unknown. In this study, we demonstrated that miR-139-5p regulates the differentiation of hematopoietic lineage cells by controlling proliferation and differentiation of hematopoietic progenitor cells. We also showed that deregulated miR-139-5p expression contributes to CML.
Materials and methods
Cell culture and transfection
HeLa and phoenix-eco cells were maintained in Dulbecco’s modified Eagle medium (Welgene) containing 10% fetal bovine serum (FBS) and 100 U/mL streptomycin and penicillin. PD31 cells were cultured in RPMI 1640 medium containing 10% FBS and 50 µM 2-mercaptoethanol. PD31 cells were transfected using a Microporator Neon system (Life Technologies). Locked nucleic acid–modified miR-139-5p inhibitor was purchased from GE Dharmacon.
Quantitative reverse transcription polymerase chain reaction (RT-PCR) and northern blot
Total RNA was isolated using TRI reagent (Molecular Research Center) according the manufacturer’s instructions. Equivalent quantities of total RNA were reverse transcribed with QuantiTect (Qiagen) and diluted complementary DNA was analyzed by real-time PCR (StepOnePlus; Applied Biosystem). Mature miR-139-5p expression was detected using TaqMan MicroRNA Assay kit (Life Technologies), and expression levels were normalized to endogenous expression of U6 small nuclear RNA. For northern blot, a total of 20 μg RNA was resolved on 15% urea-polyacrylamide gel and transferred to a membrane (Zetaprobe-GT; Bio-Rad). Antisense oligonucleotides complementary to miR-139-5p were end labeled using T4 polynucleotide kinase (Takara) and used as probe to detect miR-139-5p expression. The sequence of the primer is as follows: 5′-AGACACGTGCACTGTAGA-3′. BAS Image Analysis System (Fujifilm) was used for detection of miRNA expression.
Western blotting
Western blot was carried out as previously described.15 In brief, cells were lysed with lysis buffer [10 mM tris(hydroxymethyl)aminomethane-Cl (pH 8.5), 1 mM EDTA, 1% NP-40, 1% sodium deoxycholate, and 250 mM NaCl] supplemented with protease inhibitors (Roche). Whole cell extracts were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis gels and transferred to Immobilon-P membrane (Millipore). The immunoblot assay was performed with indicated antibodies: anti-β-actin (Sigma) and Baf60a (BD Bioscience). Antisera for Brg1 and Srg3 were raised from rabbits in our laboratory.
DNA constructs
For expression of miR-139-5p, murine miR-139-5p hairpin sequences and ∼100 nt of 5′ and 3′ flanking regions were amplified from genomic locus by PCR. The sequences of the primer pairs are as follows: 5′-GGGGACCAGCTATGGTAAGGGATGA-3′ (forward) and 5′-CTTTCTTTTGCCCACCATGCCCT-3′ (reverse). This amplicon was cloned into pGEM-T easy vector (Promega) and transferred into the pcDNA3.0 vector or MDH1-PGK-green fluorescent protein (GFP)2.0 (kindly provided by Chang-Zhen Cheng, Stanford University). For luciferase reporter assay, the fragments of Brg1 3′UTR containing the miR-139-5p binding sequences were amplified by PCR using the primers 5′-TCAGGAAGTGGCAGTGAGGAAGACTG-3′ (forward) and 5′-TCTGGTGCTACCCATCACTGCTAAGG-3′ (reverse). This fragment of Brg1 3′UTR was cloned into the pGL3_CMV vector (modified from PGL3_Baic; Promega) at the XbaI site.16 For knockdown of Brg1, target sequences (5′-CCGTCAAGGTGAAGATCAA-3′) were cloned into MDH1-PGK-GFP2.0 or murine stem cell virus (MSCV)–LTRmiR30-PIG (LMP; Open Biosystems) retroviral vectors. For knockdown of miR-139-5p, 4 consecutive miR-139-5p target sequences were cloned into MDH1-PGK-GFP2.0 vector or human nerve growth factor receptor (NGFR) vectors (GFP marker of MigR1 vector is replaced with truncated NGFR). P210BCR-ABL expression construct cloned into the NGFR retroviral vector is gift from Warren Pear (School of Medicine, University of Pennsylvania).
Mice
C57BL/6 mice were purchased from Orient Bio, maintained in specific pathogen-free barrier facilities at Seoul National University and Institute of Molecular Biology and Genetics, and used according to protocols approved by Institutional Animal Care and Use Committees of Seoul National University.
Retroviral infection and bone marrow (BM) reconstitution
The BM cells were flushed out of femurs and tibias after intraperitoneal injection with 150 mg/kg of 5-fluorouracil for 4 days. Red blood cells were removed using ammonium-chloride-potassium (ACK) lysis buffer (0.15 M NH4Cl, 10 mM KHCO3 and 0.1 mM EDTA). Then, the cells were cultured overnight in α–minimum essential medium (20% FBS, 100 U/mL penicillin, 100 U/mL streptomycin, 50 μg/mL β-mercaptoethanol, 1% nonessential amino acid [Gibco], and 1% sodium pyruvate [Gibco]) containing 5 ng/mL interleukin-3 (IL-3), 20 ng/mL IL-6, 100 ng/mL stem cell factor, and 20 ng/mL fms-like tyrosine kinase 3 (Flt3) ligand (all from Peprotech) before the initial retroviral infection. To generate retroviruses for infection into cells, the packaging phoenix-eco cells were transfected with vectors using a standard calcium phosphate protocol, and viral supernatants were harvested 2 days later. Then, cells were infected with the viral supernatants in the presence of polybrene (8 μg/mL) by spin infection for 90 minutes at 2400 rpm. This procedure was repeated 3 times, followed by the injection of 1 × 106 infected cells via the tail vein of recipient mice that are lethally γ-irradiated at 1100 rad at 2-hour intervals (137Cs, GC 3000 Elan; MDS Nordion).
Flow cytometry and cell sorting
Cells were stained with monoclonal antibodies in various combinations in 1× phosphate-buffered saline containing 1% heat-inactivated FBS. Intracelluar staining was performed according to the manufacturer’s protocols (BD Bioscience). Data were collected using FACSCanto II (BD Bioscience) and were analyzed with FlowJo software (TreeStar). FACSAria II (BD Biosciences, National Center for Inter-University Research Facilities at Seoul National University) was used for cell sorting. The antibodies used are as follows: biotin-conjugated mouse lineage (Lin) panel that contains anti-B220 (RA3-6B2), -CD3ε (145-2C11), -Gr-1 (RB6-8C5), -Mac-1 (CD11b, M1/70), -Ter-119 antibodies; CD34 (RAM34)–PE; CD117 (c-Kit, 2B8)–PE; CD127 (IL-7Rα, SB/199)–FITC; B220 (RA36B2)–APC; Mac-1 (CD11b, M1/70)–PE; and CD71 (C2)–PE (all from BD Bioscience). The following antibody conjugates were purchased from e-Biosciences: CD16/32 (FcγRII/III)–PeCy7; Sca-1 (Ly-6A/E, D7)–PE-Cy5.5; Mac-1 (M1/70)–APC eFluor 750; c-Kit (CD117, 2B8)–APC eFluor 750; and CD34 (RAM34)–eFluor 660.
Cell cycle analysis
Cell were washed twice with phosphate-buffered saline and fixed in 70% ethanol at 4°C. After that, propidium iodide (PI) staining solution (PI [Sigma] and 50 μg/mL RNase A in 0.1% bovine serum albumin) was added. Cell cycle profile was analyzed by FACSCanto II (BD Bioscience).
Luciferase reporter assay
The reporter constructs were transfected into HeLa cells using FuGENE HD (Roche). The luciferase activity in the whole cell lysates was measured with luciferase assay kit (Promega) according to the manufacturer’s instructions. The luciferase activity was normalized for transfection efficiency by β-galactosidase activity expressed from a cotransfected LacZ plasmid.
Human BM and PB samples
Fresh-frozen biopsy samples of BM and peripheral blood (PB) cells collected from normal healthy donors and patients with AML, CML, or breakpoint cluster region–Abelson–negative chronic myeloproliferative disease (CMPD) were provided from the Korean Leukemia Cell and Gene Bank. For this study, samples were rapidly thawed and subjected to RNA purification.
Statistical analysis
All statistical analyses were performed using Prism 5 (GraphPad Software, La Jolla, CA) software. Two-tailed Student t tests were used to assess the statistical significance of differences (P values) between groups. P values <.05 were considered statistically significant (*P < .05; **P < .01; ***P < .001).
Results
miR-139-5p expression is downregulated in BCR-ABL–positive CML
miR-139-5p is suppressed in several types of tumors, including hepatocellular carcinoma and gastric and breast cancers.17-19 To determine whether miR-139-5p is associated with hematopoietic malignancies, we analyzed miR-139-5p expression in the BM and PB samples from normal healthy donors and patients with several types of myeloproliferative diseases, including CML and AML. Compared with normal patients and patients with other types of leukemia, miR-139-5p was significantly downregulated in both the BM and PB of patients with chronic-phase CML (Figure 1A-B). To elucidate the relationship between CML and miR-139-5p expression, we induced a CML-like disease in mice by ectopic BCR-ABL expression, as previously described.20 Mice reconstituted with tyrosine kinase p210BCR-ABL-transduced BM cells displayed a marked increase in granulocytes and myeloid progenitor cells (supplemental Figure 1, available on the Blood Web site). Interestingly, miR-139-5p expression was significantly reduced in p210-expressing hematopoietic stem and progenitor cells (HSPCs) (Figure 1C). Moreover, miR-139-5p expression was increased in STI571 (imatinib)–treated CML cell lines (Figure 1D). To identify the signaling pathway of miR-139-5p that mediates suppression in CML, we inhibited several pathways associated with BCR-ABL signaling.21 Treatment with LY294002 (an inhibitor of the phosphatidylinositol 3-kinase [PI3K]/Akt pathway) enhanced miR-139-5p expression in CML cells, including K562 and Kcl22 cells, in contrast to the treatment with SP600125 or PD98509 (inhibitors of the JNK and MEK/extracellular signal-regulated kinase pathways, respectively) (Figure 1E; supplemental Figure 2). We also found that miR-139-5p expression was restored in CML cells by treatment with 5-aza-2′-deoxycytidine, indicating that suppression of miR-139-5p in CML is associated with DNA methylation (supplemental Figure 3). Overall, our data suggest that BCR-ABL predominantly suppressed miR-139-5p mainly through the PI3K/Akt pathway, and the low expression level of miR-139-5p was associated with CML.
The expression of miR-139-5p is downregulated in CML patients. (A) Relative expression levels of miR-139-5p in the BM were compared among samples from normal healthy donors and patients as described in the figure. Each individual dot represents 1 sample. (B) Relative miR-139-5p expression in the PB was compared among samples as described in the figure. (C) Quantitative RT-PCR analysis of miR-139-5p expression in Lin−c-Kit+ HSPCs purified from control or p210BCR-ABL-expressing mice. Data represent the mean ± standard error of the mean (SEM) (N = 4). (D) Expression of miR-139-5p in CML (K562 and Kcl22) was measured after 24-hour treatment of BCR-ABL inhibitor, STI571 (imatinib), or vehicle (dimethyl sulfoxide [DMSO]). Data represent the mean ± SEM (N = 6 for K562 cells; N = 5 for Kcl22 cells). (E) Expression of miR-139-5p in K562 cells was measured after 24-hour treatment of signaling inhibitors (LY294002 for PI3K/acutely transforming retrovirus [Akt] pathway; SP600125 for JNK pathway; PD98509 for MEK/extracellular signal-regulated kinase pathway) described in the figure. Data represent the mean ± SEM (N = 4). AML-M1, AML type 1; AML-M2, AML type 2; AML-M4, AML type 4; CML-CP, BCR-ABL–positive CML–chronic phase; CMPD-CNL, BCR-ABL–negative CMPD–chronic neutrophilic leukemia; CMPD-ET, BCR-ABL–negative CMPD–essential thrombocythemia; CMPD-MF, BCR-ABL–negative CMPD–myelofibrosis; CMPD-PV, BCR-ABL–negative CMPD–polycythemia vera.
The expression of miR-139-5p is downregulated in CML patients. (A) Relative expression levels of miR-139-5p in the BM were compared among samples from normal healthy donors and patients as described in the figure. Each individual dot represents 1 sample. (B) Relative miR-139-5p expression in the PB was compared among samples as described in the figure. (C) Quantitative RT-PCR analysis of miR-139-5p expression in Lin−c-Kit+ HSPCs purified from control or p210BCR-ABL-expressing mice. Data represent the mean ± standard error of the mean (SEM) (N = 4). (D) Expression of miR-139-5p in CML (K562 and Kcl22) was measured after 24-hour treatment of BCR-ABL inhibitor, STI571 (imatinib), or vehicle (dimethyl sulfoxide [DMSO]). Data represent the mean ± SEM (N = 6 for K562 cells; N = 5 for Kcl22 cells). (E) Expression of miR-139-5p in K562 cells was measured after 24-hour treatment of signaling inhibitors (LY294002 for PI3K/acutely transforming retrovirus [Akt] pathway; SP600125 for JNK pathway; PD98509 for MEK/extracellular signal-regulated kinase pathway) described in the figure. Data represent the mean ± SEM (N = 4). AML-M1, AML type 1; AML-M2, AML type 2; AML-M4, AML type 4; CML-CP, BCR-ABL–positive CML–chronic phase; CMPD-CNL, BCR-ABL–negative CMPD–chronic neutrophilic leukemia; CMPD-ET, BCR-ABL–negative CMPD–essential thrombocythemia; CMPD-MF, BCR-ABL–negative CMPD–myelofibrosis; CMPD-PV, BCR-ABL–negative CMPD–polycythemia vera.
Knockdown of miR-139-5p results in the expansion of mature granulocytes
To investigate the role of miRNAs in the hematopoietic system, we first examined miR-139-5p expression. We found that miR-139-5p was expressed predominantly in the BM, spleen, and lymph nodes but not in the thymus (supplemental Figure 4A). miR-139-5p expression was much higher in granulocytes than that in erythroid and B cells (supplemental Figure 4B). Furthermore, the Lin−c-Kit+Sca-1+ (LSK) compartment and granulocyte/monocyte progenitors (GMPs) expressed higher miR-139-5p levels than those of the common myeloid progenitors (CMPs) and megakaryocyte/erythrocyte progenitors (MEPs). In addition, miR-139-5p expression was positively correlated with the expression of its host gene Pde2a in hematopoietic lineage cells, suggesting that they are coexpressed (supplemental Figure 4C).
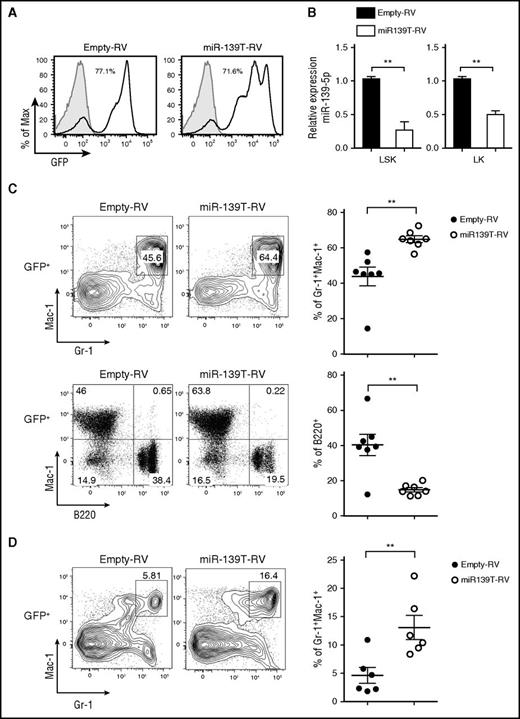
Next, to investigate the function of miR-139-5p in hematopoiesis, we knocked down miR-139-5p expression using retroviral sponge vectors expressing 4 consecutive elements, each containing the full complementary sequence to miR-139-5p (miR-139T-RV) (supplemental Figure 5).22 We transduced HSPC-enriched BM cells obtained from 5-fluorouracil-treated mice with empty-RV or miR-139T-RV and then transplanted these cells into lethally irradiated recipient mice. Two months later, we confirmed the GFP expression after transplantation (the frequency of GFP-positive cells ranged from 30% to 90% after transplantation) and knockdown of miR-139-5p in the BM cells (Figure 2A-B). Morphological inspection of the BM smear that was stained with Wright-Giemsa revealed a marked increase in mature granulocytes and exhibited dysplastic features, including irregular segmentation and hyperplasia (supplemental Figure 6). Flow cytometric analysis of GFP-positive cells in miR-139–knockdown mice revealed a significant increase in the frequency of granulocytes in the BM compared with that in control mice (Figure 2C). However, the frequency of B cells was reduced in miR-139-5p–knockdown mice. We also observed a substantial increase in granulocytes from miR-139-5p–knockdown mice (Figure 2D). Altogether, these observations indicate that the loss of miR-139-5p in the earliest stages of hematopoiesis leads to myeloid-biased differentiation.
Knockdown of miR-139-5p in HSC/progenitor causes an increase in myeloid lineage cells. (A) The BM of mice reconstituted with HSPCs transduced by empty-RV or miR-139T-RV was analyzed using GFP expression by flow cytometry. The percentage of GFP-positive cells is indicated. Gray, GFP expression in nontransduced control BM from C57BL/6 mice; black line, GFP expression in transduced BM. (B) The expression of miR-139-5p was measured in LSK and Lin−c-Kit+Sca-1− (LK) cells purified from reconstituted mice. Data represent the mean ± SEM (N = 3). (C) Representative flow cytometric analysis of granulocytes (Gr-1+Mac-1+) and B cells (B220+Mac-1−) in the BM gated on GFP-positive populations, with surface markers Gr-1, Mac-1, and B220 (upper panels). Results plotted as average frequency of granulocytes or B cells (right panels). Each individual dot represents 1 mouse. (D) Representative flow cytometric analysis of splenocytes from reconstituted mice, with Gr-1 and Mac-1 (left) and average frequency of Gr-1+Mac-1+ granulocytes (right). Each individual dot represents 1 mouse.
Knockdown of miR-139-5p in HSC/progenitor causes an increase in myeloid lineage cells. (A) The BM of mice reconstituted with HSPCs transduced by empty-RV or miR-139T-RV was analyzed using GFP expression by flow cytometry. The percentage of GFP-positive cells is indicated. Gray, GFP expression in nontransduced control BM from C57BL/6 mice; black line, GFP expression in transduced BM. (B) The expression of miR-139-5p was measured in LSK and Lin−c-Kit+Sca-1− (LK) cells purified from reconstituted mice. Data represent the mean ± SEM (N = 3). (C) Representative flow cytometric analysis of granulocytes (Gr-1+Mac-1+) and B cells (B220+Mac-1−) in the BM gated on GFP-positive populations, with surface markers Gr-1, Mac-1, and B220 (upper panels). Results plotted as average frequency of granulocytes or B cells (right panels). Each individual dot represents 1 mouse. (D) Representative flow cytometric analysis of splenocytes from reconstituted mice, with Gr-1 and Mac-1 (left) and average frequency of Gr-1+Mac-1+ granulocytes (right). Each individual dot represents 1 mouse.
miR-139-5p controls the expansion of LSK and myeloid progenitor cells
The phenotype observed in miR-139-5p–knockdown mice may be because of either alteration of the proliferation and apoptosis of granulocytes or an increase in the generation of myeloid progenitors. However, there were no significant alterations in the rate of apoptosis and proliferation in mature granulocytes (supplemental Figure 7). Thus, we investigated the effect of miR-139-5p deficiency on cells in the earliest stages of the hematopoietic process. A significant increase in myeloid progenitors, GMPs, was observed, whereas both the CMP and MEP fractions were unchanged or slightly reduced in miR-139-5p–knockdown mice, respectively (Figure 3A). We also found an ∼2.5-fold increase in the percentage of LSK in the BM of miR-139–knockdown mice (Figure 3B). Next, we investigated whether sustained expression of miR-139-5p inhibits myeloid lineage differentiation. HSPCs were transfected with retroviral vectors expressing miR-139-5p (miR-139-5p-RV) or empty-RV (Figure 4A), followed by engraftment of these cells into lethally irradiated C57BL/6 WT mouse recipients. Coexpression of GFP and miR-139-5p was detected in the BM 2 months after reconstitution (Figure 4B-C). Flow cytometric analysis of LK subsets revealed that the generation of GMPs was reduced, whereas that of CMPs was not significantly altered (Figure 4D). Sustained expression of miR-139-5p also resulted in a significant decrease in the frequency of LSK cells (Figure 4E). As ectopic expression of miR-139-5p caused an increase in MEPs, we further analyzed the effects of miR-139-5p on erythroid lineage development. Although the frequency of early erythroid precursors, including proerythroblasts (CD71hiTer-119med) and basophilic erythroblasts (CD71hiTer-119hi), was increased, that of mature red blood cells, including orthochromatophilic erythroblast cells (CD71loTer-119hi), was significantly decreased in miR-139-5p–expressing mice (supplemental Figure 8). These results indicate that ectopic expression of miR-139-5p inhibits normal erythroid maturation. Taken together, these data suggest that miR-139-5p affects HSC activity and the generation of myeloid progenitor cells during early hematopoiesis.
Knockdown of miR-139-5p results in the expansion of LSK cells and myeloid progenitors. (A) Flow cytometric analysis of hematopoietic progenitors with CD34 and FcγRIII expression in LK population (Lin−c-Kit+Sca-1−) (left). Average frequency of CMP (CD34hiFcγRIII+), GMP (CD34+FcγRIIIhi), and MEP (CD34−FcγRIII−) in LK subsets is depicted (right). (B) Representative flow cytometric profiles of LSK (Lin−c-Kit+Sca-1+) and LK (Lin−c-Kit+Sca-1−) subpopulations (left) and average frequency of LSK subsets in reconstituted mice (right).
Knockdown of miR-139-5p results in the expansion of LSK cells and myeloid progenitors. (A) Flow cytometric analysis of hematopoietic progenitors with CD34 and FcγRIII expression in LK population (Lin−c-Kit+Sca-1−) (left). Average frequency of CMP (CD34hiFcγRIII+), GMP (CD34+FcγRIIIhi), and MEP (CD34−FcγRIII−) in LK subsets is depicted (right). (B) Representative flow cytometric profiles of LSK (Lin−c-Kit+Sca-1+) and LK (Lin−c-Kit+Sca-1−) subpopulations (left) and average frequency of LSK subsets in reconstituted mice (right).
Ectopic expression of miR-139-5p suppresses the differentiation of LSK and myeloid progenitors. (A) Schematic representation of the MSCV-based retroviral vector with the insertion of miR-139-5p sequence used to overexpress miR-139-5p. The empty vector containing no insert was used as control vector. (B) Transduction efficiency was analyzed by GFP expression in the BM of reconstituted mice. (C) Expression of miR-139-5p was measured by quantitative RT-PCR in LSK and LK cells purified from reconstituted mice. Data represent the mean ± SEM (N = 3). (D) Flow cytometric analysis of hematopoietic progenitors with CD34 and FcγRIII expression (left) in LK population (Lin−c-Kit+Sca-1−). Average frequency of CMP (CD34hiFcγRIII+), GMP (CD34+FcγRIIIhi), and MEP (CD34−FcγRIII−) in LK subsets is depicted (right). (E) Representative flow cytometric profiles of LSK (Lin−c-Kit+Sca-1+) (left) and average frequency of LSK subsets in reconstituted mice (right).
Ectopic expression of miR-139-5p suppresses the differentiation of LSK and myeloid progenitors. (A) Schematic representation of the MSCV-based retroviral vector with the insertion of miR-139-5p sequence used to overexpress miR-139-5p. The empty vector containing no insert was used as control vector. (B) Transduction efficiency was analyzed by GFP expression in the BM of reconstituted mice. (C) Expression of miR-139-5p was measured by quantitative RT-PCR in LSK and LK cells purified from reconstituted mice. Data represent the mean ± SEM (N = 3). (D) Flow cytometric analysis of hematopoietic progenitors with CD34 and FcγRIII expression (left) in LK population (Lin−c-Kit+Sca-1−). Average frequency of CMP (CD34hiFcγRIII+), GMP (CD34+FcγRIIIhi), and MEP (CD34−FcγRIII−) in LK subsets is depicted (right). (E) Representative flow cytometric profiles of LSK (Lin−c-Kit+Sca-1+) (left) and average frequency of LSK subsets in reconstituted mice (right).
miR-139-5p suppresses the proliferation of CML cells and HSPCs
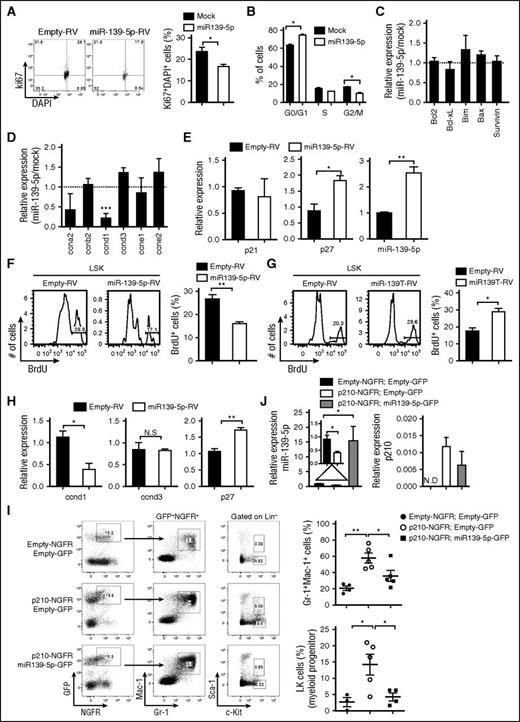
To elucidate the function of miR-139-5p in normal and malignant hematopoiesis, we first ectopically expressed miR-139-5p in CML cells. Enforced expression of miR-139-5p significantly inhibited cell proliferation via cell cycle arrest (Figure 5A-B). We also observed an increase in cell death, although there was no significant change in the expression of apoptosis-related genes, including Bcl-2 and Bcl-xL (Figure 5C; supplemental Figure 9), which may be associated with caspase signaling.23,24 However, miR-139-5p expression specifically downregulated cyclin D1, which is required in G1 in cell cycle progression (Figure 5D).25 The level of the cell cycle inhibitor p27Kip1 was increased in miR-139-5p–expressing cells, whereas p21 expression was not altered, which is consistent with a previous study13 (Figure 5E). We also found that ectopic miR-139-5p expression suppressed the proliferation of LSK and GMP cells (Figure 5F; supplemental Figure 10A). In contrast, miR-139-5p knockdown resulted in a significant increase in the frequencies of cycling LSK and GMP cells in mice (Figure 5G; supplemental Figure 10B). Similar to the CML cells, we found that cyclin D1 expression was decreased, and p27 expression was upregulated in LSK cells by miR-139-5p expression (Figure 5H). Given that miR-139-5p affects myeloid lineage differentiation and self-renewal of LSK cells, we further analyzed the transcriptional signatures in LSK cells that were transduced with miR-139-5p-RV or miR-139T-RV. The expression of stemness signatures, such as Hoxb4, Mamdc2, and Mpl, which are associated with HSC self-renewal and quiescence,26,27 was dysregulated by miR-139-5p (supplemental Figure 11). Notably, we found that miR-139-5p negatively regulated C/ebp-α and Csf1r expressions, which are essential for myeloid-lineage differentiation. Altogether, these data indicate that miR-139-5p inhibits cell proliferation by mediating cell cycle progression and suppresses myeloid differentiation by regulating the expression of transcriptional regulators during early hematopoiesis.
Expression of miR-139-5p suppresses proliferation in hematopoietic progenitors and CML cells. (A) Cell cycle profile was analyzed using Ki67/DAPI staining in Kcl22 cells after transfection with empty or miR-139-5p–expessing vectors (N = 3). (B) Cell cycle progression was analyzed using PI staining in K562 cells after transfection with empty or miR-139-5p–expessing vectors (N = 5). (C) Expression of genes that are associated with cell survival and apoptosis was analyzed in miR-139-5p–transfected K562 cells. Results are presented relative to those of cells transfected with control empty vector (N = 3). (D) Expression of cell cycle–related genes was analyzed in miR-139-5p–transfected K562 cells. Results are presented relative to those of cells transfected with control empty vector (N = 3). (E) Expression of p27, p21, and miR-139-5p in K562 cells after transfection with empty or miR-139-5p–expressing vectors was measured using quantitative RT-PCR analysis (N = 3). (F) Cell proliferation was analyzed in LSK (Lin−c-Kit+Sca-1+) of mice reconstituted with miR-139-5p-RV. Bar graph represents mean ± SEM (N = 3). (G) Cell proliferation was analyzed in LSK cells of mice reconstituted with miR-139T-RV. Bar graph represents mean ± SEM (N = 3). (H) Expression of cyclin D1, cyclin D3, and p27 was measured in LSK cells purified from mice reconstituted with miR-139-5p-RV (N = 3). (I) Representative cytometric analysis of granulocytes (left) and myeloid progenitors, LK population (middle) from reconstituted mice as described in the figure. Upper panels, empty-NGFR and empty-GFP; middle panels, p210-NGFR and empty-GFP; lower panels, p210-NGFR and miR-139-5p-GFP. The mean frequency of granulocytes and LK population is depicted (right, mean ± SEM). (J) Expression of miR-139-5p and p210 fusion transcript was analyzed in Lin−c-Kit+ HSPCs purified from reconstituted mice using quantitative RT-PCR. Data represent the mean ± SEM (N = 4).
Expression of miR-139-5p suppresses proliferation in hematopoietic progenitors and CML cells. (A) Cell cycle profile was analyzed using Ki67/DAPI staining in Kcl22 cells after transfection with empty or miR-139-5p–expessing vectors (N = 3). (B) Cell cycle progression was analyzed using PI staining in K562 cells after transfection with empty or miR-139-5p–expessing vectors (N = 5). (C) Expression of genes that are associated with cell survival and apoptosis was analyzed in miR-139-5p–transfected K562 cells. Results are presented relative to those of cells transfected with control empty vector (N = 3). (D) Expression of cell cycle–related genes was analyzed in miR-139-5p–transfected K562 cells. Results are presented relative to those of cells transfected with control empty vector (N = 3). (E) Expression of p27, p21, and miR-139-5p in K562 cells after transfection with empty or miR-139-5p–expressing vectors was measured using quantitative RT-PCR analysis (N = 3). (F) Cell proliferation was analyzed in LSK (Lin−c-Kit+Sca-1+) of mice reconstituted with miR-139-5p-RV. Bar graph represents mean ± SEM (N = 3). (G) Cell proliferation was analyzed in LSK cells of mice reconstituted with miR-139T-RV. Bar graph represents mean ± SEM (N = 3). (H) Expression of cyclin D1, cyclin D3, and p27 was measured in LSK cells purified from mice reconstituted with miR-139-5p-RV (N = 3). (I) Representative cytometric analysis of granulocytes (left) and myeloid progenitors, LK population (middle) from reconstituted mice as described in the figure. Upper panels, empty-NGFR and empty-GFP; middle panels, p210-NGFR and empty-GFP; lower panels, p210-NGFR and miR-139-5p-GFP. The mean frequency of granulocytes and LK population is depicted (right, mean ± SEM). (J) Expression of miR-139-5p and p210 fusion transcript was analyzed in Lin−c-Kit+ HSPCs purified from reconstituted mice using quantitative RT-PCR. Data represent the mean ± SEM (N = 4).
We already confirmed that miR-139-5p expression was decreased in patients with CML. In addition, dysregulated expression of genes such as p27 and cyclin D1 by miR-139-5p was associated with BCR-ABL and leukemia.28,29 Thus, we sought to determine whether the reintroduction of miR-139-5p inhibits BCR-ABL–mediated leukemogenesis. To this end, we ectopically expressed miR-139-5p in p210-transduced HSPCs, followed by engraftment of these cells into lethally irradiated C57BL/6 mice. The increased granulocyte frequency in p210-expressing mice was significantly compromised in both the BM and in the spleen by introduction of miR-139-5p (Figure 5I; supplemental Figure 12). Moreover, ectopic miR-139-5p expression negatively regulated the p210-induced expansion of myeloid progenitor cells. Expression of BCR-ABL fusion transcripts and miR-139-5p was confirmed in purified HSPCs from reconstituted mice (Figure 5J). These results indicated that ectopic miR-139-5p expression is sufficient to inhibit BCR-ABL–induced granulocytic expansion, and decreased miR-139-5p expression is closely related to hematopoietic malignancy.
Brg1 is a direct target of miR-139-5p
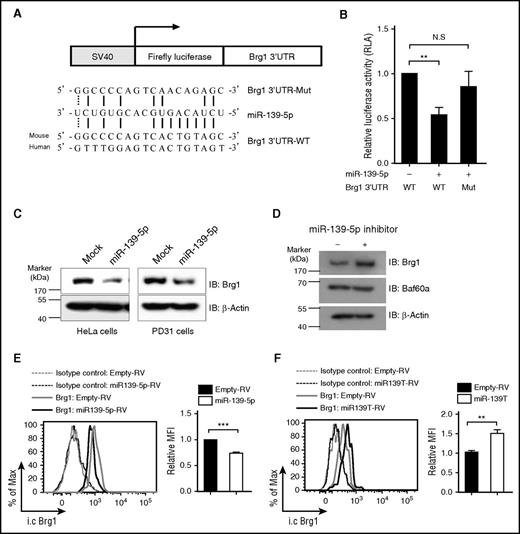
Next, we attempted to identify the direct target genes of miR-139-5p that regulate hematopoiesis. We noted that the genes for cyclin D1 and p27 did not contain the predicted binding sequence for miR-139-5p. Bioinformatics analysis using sequence-based prediction algorithms, including PicTar and TargetScan,30-32 identified Brg1, a catalytic subunit of the switch/sucrose nonfermentable (SWI/SNF) complex, as a high-scoring target gene. The Brg1-containing SWI/SNF complex has been reported to regulate cell proliferation by suppressing p27 expression and also by promoting cyclin D1 expression in diverse cell populations.33-35 To determine whether miR-139-5p directly regulates Brg1 expression, we carried out a reporter assay. We cloned the Brg1 3′UTR harboring the miR-139-5p binding sites into a luciferase reporter vector (Figure 6A). We found that ectopic miR-139-5p expression suppressed the luciferase activity (Figure 6B). However, mutations in the 5 nt of the miR-139-5p binding sites abrogated the suppression of the miR-139-5p reporter activity. Furthermore, ectopic miR-139-5p expression reduced Brg1 expression (Figure 6C). In contrast, Brg1 expression was upregulated in cells that were transfected with locked nucleic acid–modified miR-139-5p inhibitors, whereas the expression of Baf60a, another subunit of the SWI/SNF complex, was not altered (Figure 6D). We also found that Brg1 was downregulated in miR-139-5p–expressing LSK cells, whereas Brg1 expression was increased in miR-139-5p–knockdown LSK cells compared with that of control cells (Figure 6E-F). Thus, our data indicate that miR-139-5p negatively regulated Brg1 expression by binding directly to its 3′UTR.
Brg1 is a direct target of miR-139-5p. (A) Schematic representation of luciferase constructs used for reporter assay. The alignment among the binding regions of the miR-139-5p within the Brg1 3′UTR of human and mouse, mutated derivative of Brg1 3′UTR and the paring sites of miR-139-5p, and miR-139-5p sequence is shown. (B) Reporter analysis on HeLa cells transfected with constructs described in the figure. Data represent mean ± SD (N = 3). (C) western blotting of Brg1 expression in miR-139-5p–transfected HeLa (endogenous miR-139-5p expression is not detected) and PD31 (endogenous expression of miR-139-5p is detected) cells. (D) Locked nucleic acid-modified miR-139-5p inhibitor was transfected in PD31 cells, and lysates were prepared for immunoblotting with the indicated antibodies. Data are representative of 2 independent experiments. (E) Intracellular staining of Brg1 expression in LSK cells from reconstituted mice expressing miR-139-5p-RV (left). Bar graph represents relative mean fluorescence intensity (MFI) of Brg1 (N = 3, right). (F) Intracellular staining of Brg1 expression in LSK cells from miR-139-5p–knockdown mice. Bar graph represents relative MFI of Brg1 (N = 3, right).
Brg1 is a direct target of miR-139-5p. (A) Schematic representation of luciferase constructs used for reporter assay. The alignment among the binding regions of the miR-139-5p within the Brg1 3′UTR of human and mouse, mutated derivative of Brg1 3′UTR and the paring sites of miR-139-5p, and miR-139-5p sequence is shown. (B) Reporter analysis on HeLa cells transfected with constructs described in the figure. Data represent mean ± SD (N = 3). (C) western blotting of Brg1 expression in miR-139-5p–transfected HeLa (endogenous miR-139-5p expression is not detected) and PD31 (endogenous expression of miR-139-5p is detected) cells. (D) Locked nucleic acid-modified miR-139-5p inhibitor was transfected in PD31 cells, and lysates were prepared for immunoblotting with the indicated antibodies. Data are representative of 2 independent experiments. (E) Intracellular staining of Brg1 expression in LSK cells from reconstituted mice expressing miR-139-5p-RV (left). Bar graph represents relative mean fluorescence intensity (MFI) of Brg1 (N = 3, right). (F) Intracellular staining of Brg1 expression in LSK cells from miR-139-5p–knockdown mice. Bar graph represents relative MFI of Brg1 (N = 3, right).
Brg1 deficiency recapitulates the phenotypes of miR-139-5p expression
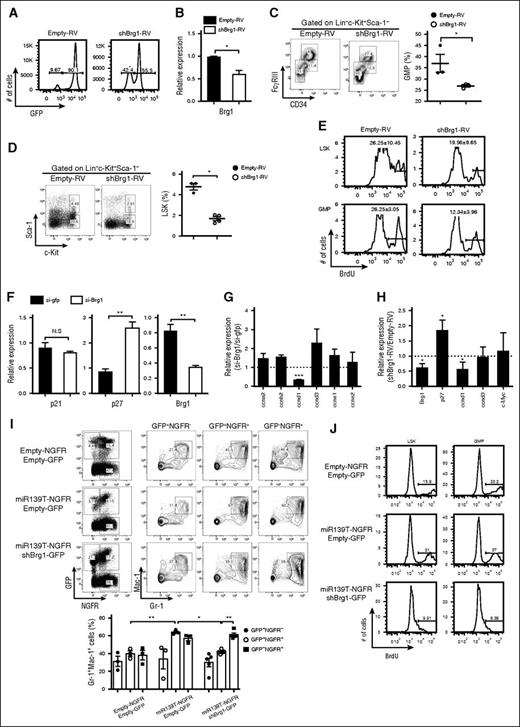
We investigated whether the phenotypes induced by miR-139-5p expression might be because of altered expression of Brg1. We also tested whether Brg1 knockdown was similar to the miR-139-5p–overexpression phenotypes. To do so, we generated Brg1-knockdown mice by reconstitution with HSPCs that were transduced with short hairpin RNA targeting Brg1 (shBrg1). The efficiencies of reconstitution and knockdown were confirmed (Figure 7A-B). Brg1 knockdown caused a reduction in myeloid progenitor cells in the BM (Figure 7C). The reduced frequency of LSK and GMP cells with a low proliferation rate in Brg1-deficient mice was similar to that observed in mice expressing ectopic miR-139-5p (Figure 7D-E). In addition, we detected a significant upregulation of p27 expression that was caused by Brg1 deletion in CML cells (Figure 7F). There was a substantial decrease in cyclin D1 transcripts, whereas other cell cycle–related genes, including cyclin A2 and cyclin D3, remained unchanged in Brg1-knockdown cells (Figure 7G). Alterations in p27 and cyclin D1 expression were also observed in Brg1-knockdown LSK cells (Figure 7H). To confirm that the increased Brg1 expression resulted in a phenotype similar to that of miR-139-5p–deficient mice, we knocked down Brg1 expression by expressing miR-139T in the BM cells and transferred the cells into lethally irradiated mice. The granulocyte frequency was significantly decreased in mice receiving miR-139T-RV together with shBrg1-RV compared with those receiving miR-139T-RV alone (Figure 7J). We also found that Brg1 depletion abolished the increase in the frequency of LSK and GMP induced by miR-139-5p knockdown (supplemental Figure 13). Furthermore, Brg1 deletion diminished the increased proliferation of LSK and GMP cells (Figure 7I). We then investigated whether Brg1 is required for BCR-ABL–mediated leukemogenesis. We knocked down Brg1 in p210-transformed mice. Brg1 depletion counteracted the increase in the frequency of granulocytes in p210-expressing mice in both the BM and in the spleen (supplemental Figure 14). Taken together, our data indicate that Brg1 is a specific target of miR-139-5p for proliferation and myeloid lineage differentiation in early hematopoiesis and for the BCR-ABL–mediated leukemogenesis induced by granulocytic expansion.
Brg1 regulates the proliferation in hematopoietic progenitors. (A) The BM of mice reconstituted with HSPCs transduced of empty-RV or shBrg1-RV was analyzed using GFP expression by flow cytometry. (B) Brg1 expression in LSK cells was measured using quantitative RT-PCR analysis (N = 3). (C) Representative profiles of lineage-committed progenitors with surface expression of CD34 and FcγRIII in LK population from Brg1-knockdown mice. Average frequency of GMP (CD34+FcγRIIIhi) in LK subsets is depicted (right). (D) Representative flow cytometric profiles of LSK (Lin−c-Kit+Sca-1+) and LK (Lin−c-Kit+Sca-1−) subpopulations (left) and average frequency of LSK subsets in Brg1-knockdown mice (right). (E) BrdU-positive cells were analyzed in LSK and GMP subsets. Data are representative of 2 independent experiments. (F) Expression of p21, p27, and Brg1 was measured in K562 cells treated with short interfering (siRNA) targeting Brg1. Data represent mean ± SEM (N = 3). (G) Expression of cell cycle–related genes was analyzed in K562 cells treated with siRNA targeting Brg1. Results are presented relative to those of cells transfected with control siRNA targeting GFP (N = 3). (H) Gene expression was analyzed in LSKs purified from Brg1-knockdown mice. Results are presented relative to those of cells transfected with control siRNA targeting GP (N = 3). (I) Representative flow cytometric analysis (upper panels) and average frequency of granulocytes (lower panels) in the BM from reconstituted mice as described in the figure. Upper panels, empty-NGFR and empty-GFP; middle panels, miR-139T-NGFR and empty-GFP; lower panels, miR-139T-NGFR and shBrg1-GFP. (J) Cell proliferation was analyzed in the subsets of LSK and GMP of reconstituted mice as described in the figure using 5-bromo-2′-deoxyuridine (BrdU) pulse labeling. Data are representative of 2 independent experiments.
Brg1 regulates the proliferation in hematopoietic progenitors. (A) The BM of mice reconstituted with HSPCs transduced of empty-RV or shBrg1-RV was analyzed using GFP expression by flow cytometry. (B) Brg1 expression in LSK cells was measured using quantitative RT-PCR analysis (N = 3). (C) Representative profiles of lineage-committed progenitors with surface expression of CD34 and FcγRIII in LK population from Brg1-knockdown mice. Average frequency of GMP (CD34+FcγRIIIhi) in LK subsets is depicted (right). (D) Representative flow cytometric profiles of LSK (Lin−c-Kit+Sca-1+) and LK (Lin−c-Kit+Sca-1−) subpopulations (left) and average frequency of LSK subsets in Brg1-knockdown mice (right). (E) BrdU-positive cells were analyzed in LSK and GMP subsets. Data are representative of 2 independent experiments. (F) Expression of p21, p27, and Brg1 was measured in K562 cells treated with short interfering (siRNA) targeting Brg1. Data represent mean ± SEM (N = 3). (G) Expression of cell cycle–related genes was analyzed in K562 cells treated with siRNA targeting Brg1. Results are presented relative to those of cells transfected with control siRNA targeting GFP (N = 3). (H) Gene expression was analyzed in LSKs purified from Brg1-knockdown mice. Results are presented relative to those of cells transfected with control siRNA targeting GP (N = 3). (I) Representative flow cytometric analysis (upper panels) and average frequency of granulocytes (lower panels) in the BM from reconstituted mice as described in the figure. Upper panels, empty-NGFR and empty-GFP; middle panels, miR-139T-NGFR and empty-GFP; lower panels, miR-139T-NGFR and shBrg1-GFP. (J) Cell proliferation was analyzed in the subsets of LSK and GMP of reconstituted mice as described in the figure using 5-bromo-2′-deoxyuridine (BrdU) pulse labeling. Data are representative of 2 independent experiments.
Discussion
In this study, we have shown that miR-139-5p was expressed in a lineage-specific manner, particularly in mature granulocytes and myeloid progenitor cells. Gain or loss of miR-139-5p in HSPCs revealed its diverse functions in hematopoiesis under steady-state conditions. Not only did miR-139-5p deficiency in hematopoietic systems promote biased differentiation into myeloid lineage with expansion of myeloid progenitor cells, but it also resulted in an aberrant proliferating capacity of hematopoietic progenitors. Downregulation of miR-139-5p was associated with human leukemia, such as BCR-ABL–induced CML. Moreover, we identified that Brg1 is a critical target of miR-139-5p during early hematopoiesis. Overall, our study provides functional evidences indicating that miR-139-5p may play a role as a key regulator in early hematopoiesis.
We demonstrated that miR-139-5p expression was strongly associated with human CML. MiR-139-5p expression was downregulated significantly in patients with CML and inhibited by the BCR-ABL oncoprotein. We found that miR-139-5p controls cell proliferation by regulating cell cycle–related genes. Ectopic miR-139-5p expression suppressed cell cycle progression by inducing p27 and inhibiting cyclin D1 expression in both LSK cells and CML cell lines. In contrast, knockdown of miR-139-5p promoted cell proliferation and caused the expansion of myeloid cells with aberrantly proliferating hematopoietic progenitor cells in the mouse model. These findings are consistent with previous studies that miR-139-5p is related to cell proliferation by regulating p27 expression and that it is associated with tumorigenesis.13,17,18,23,36-39 In addition, miR-139-5p expression was shown to be sufficient to inhibit the expansion of granulocytes that was induced by the BCR-ABL oncogene. This suggests a possibility that miR-139-5p acts as a tumor suppressor by regulating cell proliferation during early hematopoiesis.
We found that miR-139-5p specifically suppressed the expression of myeloid-affiliated C/ebp-α gene, a key regulator for granulocyte development, and the generation of GMP populations.40,41 Moreover, miR-139-5p regulates the Csf1r expression negatively in hematopoietic progenitor cells. Csf1r is essential for monocytic differentiation, and cells expressing high levels of Csf1r have a potent leukemia-initiating activity.42 These findings are consistent with a previous study showing that miR-139-5p promotes myeloid progenitor differentiation in vitro.13 Therefore, it appears that miR-139-5p regulates differentiating activity of hematopoietic progenitor cells for myeloid lineage by regulating the expression of C/ebp-α and Csf1r during early hematopoiesis.
We identified Brg1 as a functional target gene. We demonstrated that ectopic miR-139-5p expression suppressed Brg1 directly, and knockdown of this miRNA upregulated the Brg1 expression. Furthermore, Brg1 depletion inhibited the myeloid-biased expansion that was observed in miR-139-5p–knockdown mice. This appears to be achieved by negatively regulating the proliferation capacity and myeloid differentiation potential in progenitor cells. These results implicate Brg1 as a potent functional target of miR-139-5p in hematopoiesis. We also found that Brg1 is involved in the proliferation by regulating gene expressions that are affected by miR-139-5p, such as p27 and cyclin D1; this finding is consistent with previous reports that Brg1-containing SWI/SNF complex controlled p27 and cyclin D1 expression and was required for cell proliferation of HSCs.33-35 It is also known that Brg1-containing SWI/SNF complex regulates myeloid lineage differentiation by promoting C/ebp-α expression in HSCs and directly controls Csf1r transcripts by binding to its distal regulatory locus.35,43 Our results consistently demonstrated that Brg1 directed myeloid differentiation by regulating C/ebp-α and Csf1r expressions in LSK cells. Along with these findings, our evidences suggest that Brg1 is a key target gene of miR-139-5p involved in proliferation and differentiation of myeloid cells, and deregulation of Brg1 during early hematopoiesis may contribute to leukemogenesis. This hypothesis is consistent with previous reports that Brg1-containing SWI/SNF complex was essential for leukemia maintenance.35,44,45 In addition, increasing evidences have demonstrated that Brg1 is expressed at high levels in several types of primary cancers and interacts with oncoproteins, including c-Myc, MITF, and β-catenin to promote their survival and proliferation.46-51 Further studies are needed to reveal a precise mechanism for the Brg1-mediated initiation of leukemogenesis.
In summary, our data collectively indicate that miR-139-5p is a critical regulator for cell proliferation during early hematopoiesis. MiR-139-5p functions to fine-tune the Brg1 expression. Interestingly, changes in miR-139-5p expression are associated with hematopoietic malignancies such as CML, and miR-139-5p expression resulted in the remission of CML-like disease. Thus, miR-139-5p may serve as a potential therapeutic target for hematopoietic malignancies.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the Korea Leukemia Bank for providing the patient samples.
This work was supported by grants from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute, funded by the Ministry of Health and Welfare, Republic of Korea (HI14C0311), and the Korea Mouse Phenotyping Project (2014M3A9D5A01073789) and Basic Science Research program (2016R1A2B3013865) through the National Research Foundation of Korea (NRF), funded by the Ministry of Science, Global Information & Communication Technology, and Future Planning. J.C. and K.P. are supported by the BK21 Plus program (F14SN01D1305) from the Ministry of Education and NRF, Korea. J.C. is supported by Seoul Science Fellowship.
Authorship
Contribution: J.C. and R.H.S. conceived the project, designed the experimental approach, interpreted data, and wrote the manuscript; Y.-K.K., V.N.K., K.P., and J.N. helped with analysis of some experiments and provided technical assistance; S.-S.Y. provided imatinib and helped with analysis of CML cells; and D.-W.K. provided human samples and helped with analysis of some experiments.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for Y.-K.K. is Department of Biochemistry, Chonnam National University Medical School, Gwangju, Korea.
Correspondence: Rho Hyun Seong, School of Biological Sciences and Institute of Molecular Biology and Genetics, Seoul National University, Gwanak-gu, Gwanak-ro 599, Seoul 151-742, Korea; e-mail: rhseong@snu.ac.kr.

![Figure 1. The expression of miR-139-5p is downregulated in CML patients. (A) Relative expression levels of miR-139-5p in the BM were compared among samples from normal healthy donors and patients as described in the figure. Each individual dot represents 1 sample. (B) Relative miR-139-5p expression in the PB was compared among samples as described in the figure. (C) Quantitative RT-PCR analysis of miR-139-5p expression in Lin−c-Kit+ HSPCs purified from control or p210BCR-ABL-expressing mice. Data represent the mean ± standard error of the mean (SEM) (N = 4). (D) Expression of miR-139-5p in CML (K562 and Kcl22) was measured after 24-hour treatment of BCR-ABL inhibitor, STI571 (imatinib), or vehicle (dimethyl sulfoxide [DMSO]). Data represent the mean ± SEM (N = 6 for K562 cells; N = 5 for Kcl22 cells). (E) Expression of miR-139-5p in K562 cells was measured after 24-hour treatment of signaling inhibitors (LY294002 for PI3K/acutely transforming retrovirus [Akt] pathway; SP600125 for JNK pathway; PD98509 for MEK/extracellular signal-regulated kinase pathway) described in the figure. Data represent the mean ± SEM (N = 4). AML-M1, AML type 1; AML-M2, AML type 2; AML-M4, AML type 4; CML-CP, BCR-ABL–positive CML–chronic phase; CMPD-CNL, BCR-ABL–negative CMPD–chronic neutrophilic leukemia; CMPD-ET, BCR-ABL–negative CMPD–essential thrombocythemia; CMPD-MF, BCR-ABL–negative CMPD–myelofibrosis; CMPD-PV, BCR-ABL–negative CMPD–polycythemia vera.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/128/17/10.1182_blood-2016-02-702464/4/m_2117f1.jpeg?Expires=1769083232&Signature=e1mC6YU77ljSPqmI9iSA5-lyCyzChE4MjpdyaMxFQfMtaKT9PiBaO55ZM6F6HNniNvD4e3rlmJ4SVQneJIoaAXpLJSf4FDLgolHdEqHnGwN4tALORXvP7HYDMLKHpYYBeKA08-vmaeEJprugjZ~dDxEbHWIce9RqcwQKMMhfETCr4NBboj6kgWtlB2QDdvV9CN3GCQm6DYEjsh6tcI54j71RqZLJRjz1xBhwFMOKdIqk2W7BHFffzi7tuSIO29fxfJp0yQgC3pThYYhnUrs9WEq-ZebA9beywSc0iNF8C-ZUYnRL07Q5jQpQvPcqw91gJ-RkMuTDN6uXCyMZxhpFAw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal