Key Points
Only a minority of myeloma cell lines are killed when the prosurvival BCL2 or BCLXL are selectively inhibited with BH3 mimetic compounds.
In contrast, targeting MCL1 readily killed ∼70% of the myeloma cell lines tested, including both low-passage and well-established ones.
Abstract
New therapeutic targets are needed to address the poor prognosis of patients with high-risk multiple myeloma. Myeloma cells usually express a range of the prosurvival BCL2 proteins. To define the hierarchy of their relative importance for maintaining the survival of myeloma cells, we targeted each of them in a large panel of cell lines, using pharmacological inhibitors or gene editing or by peptide-based approaches, alone or in combination. The majority of well-established immortalized cell lines (17/25) or low-passage myeloma cell lines (5/7) are readily killed when MCL1 is targeted, even including those cell lines sensitive to BCL2 inhibition. Targeting MCL1 also constrained the growth of myeloma in vivo. We also identified a previously unrecognized subset of myeloma that is highly BCLXL-dependent, and has the potential for cotargeting MCL1 and BCLXL. As MCL1 is pivotal for maintaining survival of most myelomas, it should be prioritized for targeting in the clinic once high-quality, validated inhibitors become available.
Introduction
Multiple myeloma is a malignancy of antibody-secreting plasma cells.1 Despite recent advances in our understanding of the genetic basis of this disease and the introduction of novel therapies, the outcome for many patients remains poor.2,3 For example, 20% of newly diagnosed patients with stage 3 myeloma have disease marked by high lactate dehydrogenase levels or unfavorable cytogenetic abnormalities (eg, 17p deletion, t[4;14]). Half these patients die within 2 years of diagnosis, in spite of advanced therapies. Hemizygous loss of the tumor suppressor TP53, typically because of the deletion of chromosome 17p, with or without concomitant loss of protein expression4 or mutation of the remaining allele,5 is associated with a poor prognosis.6,7 In such patients, responses to targeted therapies such as immune modulator agents remain suboptimal, and allogeneic stem cell transplantation, with its associated risks, remains the only curative treatment. Hence, there is great interest in discovering and developing novel therapeutic approaches for this disease.2,3
Our focus is on targeting the BCL2-regulated cell survival pathway, also known as the mitochondrial or intrinsic cell survival pathway.8 Results from early-phase clinical trials with ABT-263 (navitoclax),9 and more recently ABT-199 (venetoclax),10 reveal that the prosurvival protein BCL2 is a highly promising target for treating some types of B-cell malignancies. About 80% of patients with refractory or relapsed chronic lymphocytic leukemia respond to ABT-199. In some, their responses have been dramatic and durable. Interestingly, studies using myeloma cell lines have suggested that the targeting BCL2 holds promise for patients with multiple myeloma.11,12 However, in a clinical trial of ABT-199 in patients with heavily pretreated myeloma, objective responses were observed in only 2, both t(11;14), of 32 evaluable patients, suggesting only a minor subset of myeloma is BCL2-dependent.13,14
We set out to determine which prosurvival BCL2 proteins are responsible for the survival of a panel of myeloma cell lines, in anticipation that our findings are likely to be highly relevant to patients with multiple myeloma. Normally, BCL2 and its prosurvival relatives such as BCLXL and MCL1 maintain cellular viability by restraining the activity of cell death mediators BAX and BAK.15 When a cell is no longer required, stressed, or damaged, these prosurvival proteins are inhibited by the BH3-only proteins (eg, BIM), thus allowing BAX and BAK to drive mitochondrial outer membrane permeabilization.16 ABT-199 selectively inhibits BCL2, but not its other prosurvival relatives, thereby triggering apoptosis in cells that rely on BCL2 for their survival.17 It mimics the action of the killer BH3-only proteins; hence, this class of small molecule inhibitors is known as the BH3 mimetics.18
By pharmacologically inhibiting the prosurvival BCL2 proteins, we found that a fraction of the myeloma cell lines were rapidly killed when BCL2 (25%) or BCLXL (25%) were targeted with small-molecule inhibitors. In contrast, the majority (∼70%) of the cell lines tested are heavily reliant on MCL1. Moreover, targeting MCL1 halted the otherwise inevitable progression of this disease when tumor cells are inoculated into mice. Our studies lend strong support to the notion that the MCL1 is a high-value therapeutic target for advancing the treatment of multiple myeloma, particularly for those patients with characteristics that currently carry a poor prognosis.
Methods
Cell lines
All myeloma cell lines used are described in supplemental Table 1, available on the Blood Web site. The low-passage patient-derived myeloma cell lines ALF-1/TK-2 and ALF-2/TK-1 have been described.19 Using similar approaches, additional low-passage lines (TK-3 to TK-8) were generated and used in this study. BAX−/−BAK−/− cells were generated using CRISPR/Cas9.
Reagents
ABT-737, ABT-199, and A-1155463 were provided by AbbVie; A-1210477 (Active Biochem), obatoclax (Selleck), doxycycline (Sigma), and Q-VD-OPH (MP Biomedicals) were purchased. A-1331852 was synthesized as previously described.20
Plasmids
CRISPR/Cas9 gene editing
Myeloma cells were serially infected with lentiviruses expressing Cas9 mCherry and guide RNAs (sgRNA) GFP. To induce expression of the sgRNA, doxycycline was added to tissue culture medium at a final concentration of 1 μg/mL. After 72 hours, cell lysates were prepared for immunoblotting. In addition, genomic DNA was sequenced to confirm the mutation of targeted DNA by using the Illumina MiSeq platform.22 For sequencing, PCR primers with overhang sequences for each sgRNA are described (supplemental Table 3).
Statistics
GraphPad Software was used for statistical analysis. All data are expressed as means ± SD and analyzed with an unpaired Student t test for statistical significance. P values < .05 were considered to be significant.
Study approvals
All animal experiments were approved by the Walter and Eliza Hall Institute Animal Ethics Committee and conducted according to its guidelines. Written informed consents were obtained from patients for the collection of bone marrow samples. The studies were performed with the approval of the Alfred Hospital’s Human Research Ethics Committee.
Methods for lentivirus production and infection, drug sensitivity of primary myeloma cells, cell viability assays, immunoblotting, in vivo imaging, and histology are provided as supplemental data.
Results
Treating human myeloma cell lines with validated BH3 mimetic compounds
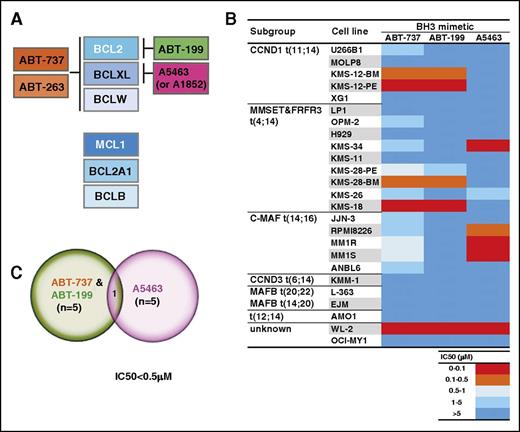
To determine whether human myeloma cell lines rely on 1 or more of the prosurvival BCL2 proteins for their survival, we treated a panel of 25 well-established myeloma cell lines with validated inhibitors of these proteins (Figure 1A; experimental strategy is outlined in supplemental Figure 1). ABT-737 targets BCL2, BCLXL, and BCLW,23,24 whereas ABT-199 has significantly lower affinity for BCLXL and BCLW.17 In contrast, 2 recently described compounds, A-1155463 and A-1331852 (hereafter, referred to as A5463 and A1852), act to target BCLXL selectively20,25 ; they have no appreciable affinity for the other prosurvival BCL2 proteins.
A subset of human myeloma cell lines are readily killed when BCL2 or BCLXL is inhibited with small molecules. (A) Selectivity of well-validated BH3 mimetic compounds for the prosurvival BCL2 proteins. Of the 6 human prosurvival BCL2 proteins, ABT-737 selectively targets BCL2, BCLXL and BCLW.23,24 ABT-263 (navitoclax) is a clinical candidate compound in the same chemical class as ABT-737, and has identical target selectivity.51 The BCL2 selective inhibitor ABT-199 (venetoclax) was developed by modifying ABT-263 such that its affinity for BCLXL and BCLW is reduced.17 Although ABT-737, ABT-263, and ABT-199 are related in their chemical structures, the BCLXL-selective inhibitors A-1155463 and A-1331852 (referred to as A5463 and A1852) belong to a structurally distinct class.20,25 (B) Sensitivity of diverse human myeloma cell lines to the BH3 mimetics. The sensitivity of the 25 myeloma cell lines (details provided in supplemental Table 1) to ABT-737, ABT-199, or A5463 was determined after culturing in 0 to 10 μM of the indicated compound for 48 hours. Cell viability was determined using the CellTiter-Glo assay. IC50 values were calculated using data from 3 independent experiments (eg, in supplemental Figure 1). A discrete heat map representation of the mean IC50s is shown in this panel (actual values are shown in supplemental Table 4): red represents potent killing (IC50 < 0.1 μM), whereas blue indicates resistance (IC50 > 5 μM). (C) Sensitivity of the multiple myeloma cell lines to the selective inhibition of BCL2 or BCLXL. Venn diagram summarizing the sensitivity of the cell lines tested to ABT-737, ABT-199, or A5463. Five lines (KMS-12-BM, KMS-12-PE, KMS-28-BM, KMS-18, and WL-2) are highly sensitive (arbitrarily defined as IC50 < 0.5 μM) to ABT-737, and in agreement with previous studies, the same lines are also very responsive to the BCL2-selective inhibitor ABT-199.11,12 The selective inhibition of BCLXL with A5463 is efficacious in a subset of 5 cell lines: 4 (KMS-34, RPMI-8226, MM1R, MM1S) are distinct from the ABT-737/ABT-199 sensitive ones.
A subset of human myeloma cell lines are readily killed when BCL2 or BCLXL is inhibited with small molecules. (A) Selectivity of well-validated BH3 mimetic compounds for the prosurvival BCL2 proteins. Of the 6 human prosurvival BCL2 proteins, ABT-737 selectively targets BCL2, BCLXL and BCLW.23,24 ABT-263 (navitoclax) is a clinical candidate compound in the same chemical class as ABT-737, and has identical target selectivity.51 The BCL2 selective inhibitor ABT-199 (venetoclax) was developed by modifying ABT-263 such that its affinity for BCLXL and BCLW is reduced.17 Although ABT-737, ABT-263, and ABT-199 are related in their chemical structures, the BCLXL-selective inhibitors A-1155463 and A-1331852 (referred to as A5463 and A1852) belong to a structurally distinct class.20,25 (B) Sensitivity of diverse human myeloma cell lines to the BH3 mimetics. The sensitivity of the 25 myeloma cell lines (details provided in supplemental Table 1) to ABT-737, ABT-199, or A5463 was determined after culturing in 0 to 10 μM of the indicated compound for 48 hours. Cell viability was determined using the CellTiter-Glo assay. IC50 values were calculated using data from 3 independent experiments (eg, in supplemental Figure 1). A discrete heat map representation of the mean IC50s is shown in this panel (actual values are shown in supplemental Table 4): red represents potent killing (IC50 < 0.1 μM), whereas blue indicates resistance (IC50 > 5 μM). (C) Sensitivity of the multiple myeloma cell lines to the selective inhibition of BCL2 or BCLXL. Venn diagram summarizing the sensitivity of the cell lines tested to ABT-737, ABT-199, or A5463. Five lines (KMS-12-BM, KMS-12-PE, KMS-28-BM, KMS-18, and WL-2) are highly sensitive (arbitrarily defined as IC50 < 0.5 μM) to ABT-737, and in agreement with previous studies, the same lines are also very responsive to the BCL2-selective inhibitor ABT-199.11,12 The selective inhibition of BCLXL with A5463 is efficacious in a subset of 5 cell lines: 4 (KMS-34, RPMI-8226, MM1R, MM1S) are distinct from the ABT-737/ABT-199 sensitive ones.
Of the 25 lines tested, 5 (20%) were highly sensitive (defined as IC50 < 0.5 μM) to ABT-737 (Figure 1B; supplemental Table 4). Interestingly, these same lines were also very sensitive to killing induced by the BCL2 inhibitor ABT-199. This strongly suggested that the selective targeting of BCL2 alone was sufficient to kill these 5 cell lines. Of note, 2 of these 5 lines (KMS-12-BM, KMS-12-PE) harbored the t(11;14) chromosomal translocation.
Our finding that 20% of the myeloma cell lines rely on BCL2 is in good general agreement with previously reported studies.11,12 Our data also suggest that previously unrecognized cases of myeloma, such as those harboring the t(4;14) chromosomal translocation (eg, KMS-28-BM, KMS-18), are also likely to be promising candidates for therapy with a BCL2 inhibitor. Having confirmed the role for BCL2 in myeloma, we next asked whether BCLXL might also have a role.
A distinct subset of myeloma cell lines are readily killed when BCLXL is targeted
Although ABT-737 can inhibit BCLXL, our previous functional studies had indicated that it is a relatively ineffective BCLXL inhibitor in lymphoid cells.26 Given this caveat, and to fully define the role of BCLXL in myeloma, we used a novel and highly potent inhibitor of BCLXL.20,25,27 Importantly, A5463 is structurally distinct from ABT-737 and ABT-199.
We found that, of the 25 cell lines, 5 (20%) of them were readily killed by A5463. Of these, only 1 (WL-2) was also readily killed by ABT-737 (and ABT-199; Figure 1B). In contrast, 4 other cell lines highly sensitive to A5463 (KMS-34, RPMI-8226, MM1R, MM1S) were relatively insensitive to ABT-737 (10-80-fold weaker) and completely resistant to ABT-199. In summary, our initial screens identified subsets of human myeloma cell lines that rely on BCL2, as previously described,11,12 or interestingly, on BCLXL2,7 with very little overlap between these subsets (Figure 1C). Although all sensitive cells prominently expressed the target (BCL2 or BCLXL), high expression level was not predictive of sensitivity (supplemental Figure 2).
Our data suggest that BCLXL on its own should be considered a potential therapeutic target in multiple myeloma, in addition to BCL2. It is also noteworthy that suboptimal small molecule inhibitors may not fully reveal the full effect of targeting specific prosurvival proteins, as is the case with using ABT-737 to functionally inhibit BCLXL (Figure 1B). Our data strongly suggest that studies to test for sensitivity to BCLXL inhibition are best conducted using a selective BCLXL inhibitor, such as A5463, not just with ABT-737, as the latter does not appear to be as potent at inhibiting BCLXL as A5463 (supplemental Figure 3).26
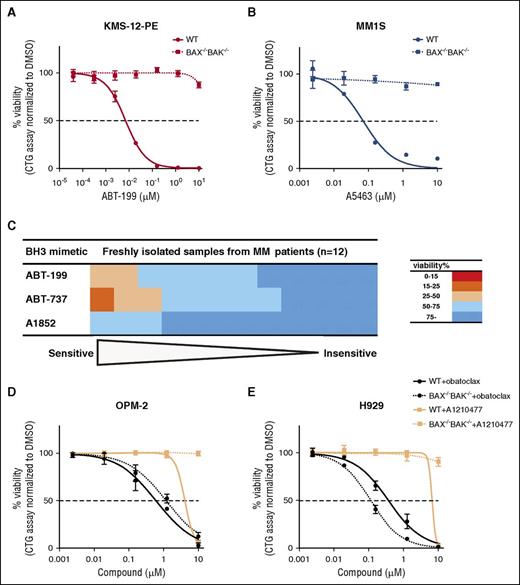
Having established the potency of A5463, we also formally tested whether it acts specifically to induce apoptosis (Figure 2; supplemental Figures 4-5). In mechanistic studies using KMS-12-PE or MM1S cells, cell lines sensitive to BCL2 or BCLXL inhibition, respectively, we found that the absence of the downstream mediators of apoptosis, BAX and BAK (Figure 2A-B; supplemental Figure 4), completely abolished killing by these BH3 mimetics, thereby precluding any off-target or nonspecific cytotoxic effects these compounds might harbor. We also established that A5463 killed MM1S cells by a caspase-dependent mechanism (supplemental Figure 5C), whereas this compound had negligible effect on the viability of KMS-12-PE cells, a line sensitive to BCL2 inhibition.
Potency and specificity of BH3 mimetic compounds in myeloma cell lines or primary myeloma samples. (A) BAX/BAK-dependent killing of KMS-12-PE myeloma cells by an inhibitor of BCL2. The viability of WT KMS-12-PE cells or ones engineered to lack BAX and BAK 48 hours after treatment with 0 to 10 μM of the BCL2-selective inhibitor ABT-199 was determined using the CellTiter-Glo assay. (B) Similar results to A, but obtained from treating WT or BAX−/−BAK−/− MM1S myeloma cells with the BCLXL inhibitor A5463. (C) Sensitivity of primary myeloma samples to the BH3 mimetics. The viability of 12 primary myeloma samples were determined 72 h after treatment with 1 μM ABT-199, ABT-737, or the BCLXL inhibitor A1852 ex vivo; the samples were ranked according to their sensitivity to each drug. (D) Sensitivity of a MCL1-dependent cell line OPM-230 to putative inhibitors of MCL1. The viability of WT OPM-2 or a BAX−/−BAK−/−-deficient clone 48 hours after treatment with 0 to 10 μM of obatoclax (in black) or the MCL1-selective inhibitor A-1210477 (in blue) was determined using the CellTiter-Glo assay. (E) Sensitivity of another MCL1-dependent cell line H92933,35 to putative inhibitors of MCL1. Similar experiments to that described in panel D, but undertaken with H929 cells instead. The dotted horizontal line represents 50% loss in viability. The results in panels A, B, D, and E represent the mean values from 3 independent experiments; data are shown as means ± 1 SD.
Potency and specificity of BH3 mimetic compounds in myeloma cell lines or primary myeloma samples. (A) BAX/BAK-dependent killing of KMS-12-PE myeloma cells by an inhibitor of BCL2. The viability of WT KMS-12-PE cells or ones engineered to lack BAX and BAK 48 hours after treatment with 0 to 10 μM of the BCL2-selective inhibitor ABT-199 was determined using the CellTiter-Glo assay. (B) Similar results to A, but obtained from treating WT or BAX−/−BAK−/− MM1S myeloma cells with the BCLXL inhibitor A5463. (C) Sensitivity of primary myeloma samples to the BH3 mimetics. The viability of 12 primary myeloma samples were determined 72 h after treatment with 1 μM ABT-199, ABT-737, or the BCLXL inhibitor A1852 ex vivo; the samples were ranked according to their sensitivity to each drug. (D) Sensitivity of a MCL1-dependent cell line OPM-230 to putative inhibitors of MCL1. The viability of WT OPM-2 or a BAX−/−BAK−/−-deficient clone 48 hours after treatment with 0 to 10 μM of obatoclax (in black) or the MCL1-selective inhibitor A-1210477 (in blue) was determined using the CellTiter-Glo assay. (E) Sensitivity of another MCL1-dependent cell line H92933,35 to putative inhibitors of MCL1. Similar experiments to that described in panel D, but undertaken with H929 cells instead. The dotted horizontal line represents 50% loss in viability. The results in panels A, B, D, and E represent the mean values from 3 independent experiments; data are shown as means ± 1 SD.
As only a fraction of the myeloma cell line panel tested was killed by inhibition of BCL2 or BCLXL, we next asked whether this conclusion is borne out in samples freshly isolated from patients with multiple myeloma (Figure 2C). Consistent with our cell line data, we found that a similarly minor fraction of these primary patient-derived samples were killed by ABT-199 treatment. Our data indicate that the majority (75%, 9/12 patient samples) were refractory to the selective inhibition of BCL2 or BCLXL, or when both of these prosurvival proteins are targeted.
On the basis of these studies, we thus conclude that only some patients with myeloma are likely to respond to ABT-199, and that inhibition of BCLXL on its own may have a role for treating multiple myeloma.
Testing for MCL1 dependence with small molecule inhibitors
We then asked which other prosurvival BCL2 protein is responsible for keeping the other myeloma cell lines alive. A prime candidate is MCL1, as normal plasma cells rely on it28,29 and previous studies had implicated MCL1 in myeloma.30,31 We tested 2 recently described MCL1 inhibitors, obatoclax32 and A-1210477,33,34 for their cytotoxic activity in 2 myeloma cell lines that depend on MCL1; namely, OPM-230 and H929.35
We readily discounted obatoclax,32 as it killed both the parental wild-type (WT) cells as well as ones engineered to be devoid of BAX and BAK (Figure 2D-E). This is consistent with the notion that the cytotoxic action of obatoclax cannot be solely accounted for by inducing apoptosis.36,37 In contrast, A-121047733,34 appeared much more promising, as it acted selectively (Figure 2D-E). However, its lack of potency (IC50 > 5 μM), unlike that of ABT-199 or A5463 (Figure 2A-B), made it a less than optimal tool compound for our myeloma studies. Thus, neither obatoclax nor A-1210477 is optimal for our attempts to clearly define the role of MCL1 in myeloma.
Genetically targeting the prosurvival BCL2 proteins, using CRISPR/Cas9 genome editing technology
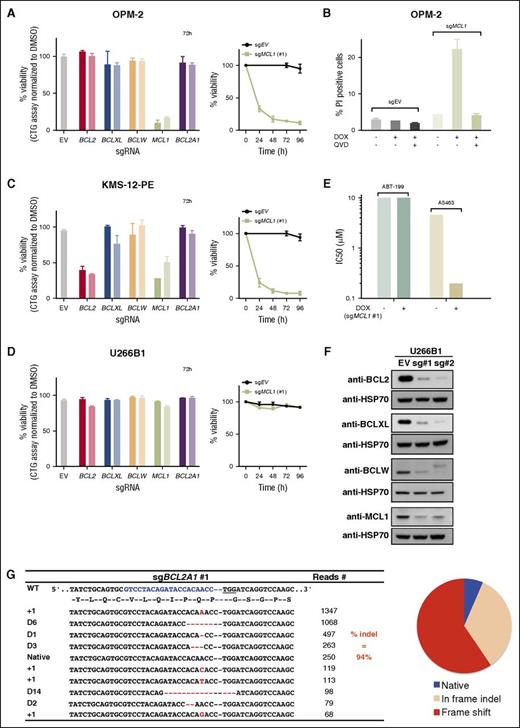
As there are no validated small molecules suitable to specifically and efficiently inhibit MCL1, we next investigated the feasibility of alternative approaches to address the question of what maintains their survival. Initially, we turned to targeting the expression of the pro-survival BCL2 proteins, using CRISPR/Cas9 genome editing technology.38,39 Using OPM-2 cells as a MCL1-dependent model cell line,30 we evaluated the effect of inactivating BCL2, BCLXL, BCLW, MCL1, or BCL2A1 in this cell line when expression of sgRNAs targeting these genes was induced (Figure 3A).22
Efficient CRISPR/Cas9-mediated gene editing to mutate genes that encode the prosurvival BCL2 proteins. (A) The OPM-2 cell line relies on MCL1 for its survival. The viability of OPM-2 cells 72 hours after addition of doxycycline (DOX) to induce expression of sgRNAs that target BCL2, BCLXL, BCLW, MCL1, or BCL2A1 was determined with CellTiter-Glo assays.22 Two sgRNAs were tested for each gene: sgRNA#1 (darker shade) and sg#2 (lighter shade). The panel on the right shows the viability of cells expressing the empty vector (EV; control) or MCL1 sgRNA#1 0 to 96 hours after addition of DOX to induce expression of the guide RNAs. (B) Caspases mediate the killing of OPM-2 cells when MCL1 is genetically targeted. The proportion of dying cells (PI uptake determined by flow cytometry) of OPM-2 cells 24 hours after switching on the expression of the sgRNA (+ DOX) to target MCL1 was determined when cultured with (+) or without (−) the broad-spectrum caspase inhibitor Q-VD-OPh (25 μM). Data (means ± 1 SD) are derived from 2 independent experiments. (C) KMS-12-PE cells rapidly lose their viability when BCL2 or MCL1 is genetically ablated. Similar experiments to that in A, but undertaken with KMS-12-PE cells. Although genetically targeting BCLXL, BCLW, or BCL2A1 has no effect, targeting BCL2 or MCL1 readily kills KMS-12-PE cells. (D) Deleting BCL2, BCLXL, BCLW, MCL1, or BCL2A1 had no effect on the viability of U266B1 cells. Similar experiments to that in A and C, but undertaken with U266B1 cells. (E) U266B1 cells rely on MCL1 and BCLXL for their survival. The sensitivity (in IC50s) of U266B1 cells to selective inhibition of BCL2 (with ABT-199) or BCLXL (with A5463) was determined at 48 hours when MCL1 was genetically targeted (+DOX) or not (−DOX). Of note, the concomitant targeting of BCLXL (pharmacologically) and MCL1 (genetically) readily killed U266B1 cells, whereas the cotargeting of BCL2 and MCL1 did not. (F) Ablating BCL2, BCLXL, BCLW, or MCL1 protein expression. Immunoblotting of cell lysates prepared from pools of the human myeloma cell line U266B1 after DOX addition for 72 hours to induce expression of sgRNAs that target expression of the indicated proteins. Compared with the control cells expressing the empty vector (EV; left lanes), the levels of BCL2, BCLXL, BCLW, or MCL1 are significantly reduced in the pools analyzed. HSP70, loading control. (G) Efficient deletion of BCL2A1. Genomic DNA prepared from U266B1 expressing an sgRNA to BCL2A1 (sgRNA #1) was subjected to Sanger DNA sequencing. The table shows the top 10 sequencing reads; 94% of all the reads (3902 in total) were indels. Of these, the majority is predicted to be deleterious to expression of BCL2A1 by causing frame shift mutations (red in the pie chart); some were in frame indels (beige). Red dashes, deleted bases; red letters, insertions or substitutions. The sequencing data for all the sgRNAs used in our study, including those of the cells shown in panel D, as well as their characterization, are shown in supplemental Figure 5-13. The results in panels A, C, D, and E represent the data from representative experiments performed in triplicates and shown as means ± 1 SD.
Efficient CRISPR/Cas9-mediated gene editing to mutate genes that encode the prosurvival BCL2 proteins. (A) The OPM-2 cell line relies on MCL1 for its survival. The viability of OPM-2 cells 72 hours after addition of doxycycline (DOX) to induce expression of sgRNAs that target BCL2, BCLXL, BCLW, MCL1, or BCL2A1 was determined with CellTiter-Glo assays.22 Two sgRNAs were tested for each gene: sgRNA#1 (darker shade) and sg#2 (lighter shade). The panel on the right shows the viability of cells expressing the empty vector (EV; control) or MCL1 sgRNA#1 0 to 96 hours after addition of DOX to induce expression of the guide RNAs. (B) Caspases mediate the killing of OPM-2 cells when MCL1 is genetically targeted. The proportion of dying cells (PI uptake determined by flow cytometry) of OPM-2 cells 24 hours after switching on the expression of the sgRNA (+ DOX) to target MCL1 was determined when cultured with (+) or without (−) the broad-spectrum caspase inhibitor Q-VD-OPh (25 μM). Data (means ± 1 SD) are derived from 2 independent experiments. (C) KMS-12-PE cells rapidly lose their viability when BCL2 or MCL1 is genetically ablated. Similar experiments to that in A, but undertaken with KMS-12-PE cells. Although genetically targeting BCLXL, BCLW, or BCL2A1 has no effect, targeting BCL2 or MCL1 readily kills KMS-12-PE cells. (D) Deleting BCL2, BCLXL, BCLW, MCL1, or BCL2A1 had no effect on the viability of U266B1 cells. Similar experiments to that in A and C, but undertaken with U266B1 cells. (E) U266B1 cells rely on MCL1 and BCLXL for their survival. The sensitivity (in IC50s) of U266B1 cells to selective inhibition of BCL2 (with ABT-199) or BCLXL (with A5463) was determined at 48 hours when MCL1 was genetically targeted (+DOX) or not (−DOX). Of note, the concomitant targeting of BCLXL (pharmacologically) and MCL1 (genetically) readily killed U266B1 cells, whereas the cotargeting of BCL2 and MCL1 did not. (F) Ablating BCL2, BCLXL, BCLW, or MCL1 protein expression. Immunoblotting of cell lysates prepared from pools of the human myeloma cell line U266B1 after DOX addition for 72 hours to induce expression of sgRNAs that target expression of the indicated proteins. Compared with the control cells expressing the empty vector (EV; left lanes), the levels of BCL2, BCLXL, BCLW, or MCL1 are significantly reduced in the pools analyzed. HSP70, loading control. (G) Efficient deletion of BCL2A1. Genomic DNA prepared from U266B1 expressing an sgRNA to BCL2A1 (sgRNA #1) was subjected to Sanger DNA sequencing. The table shows the top 10 sequencing reads; 94% of all the reads (3902 in total) were indels. Of these, the majority is predicted to be deleterious to expression of BCL2A1 by causing frame shift mutations (red in the pie chart); some were in frame indels (beige). Red dashes, deleted bases; red letters, insertions or substitutions. The sequencing data for all the sgRNAs used in our study, including those of the cells shown in panel D, as well as their characterization, are shown in supplemental Figure 5-13. The results in panels A, C, D, and E represent the data from representative experiments performed in triplicates and shown as means ± 1 SD.
Interestingly, inducing the expression of sgRNAs to MCL1 in OPM-2 cells led to the rapid loss of cell viability, whereas targeting other prosurvival BCL2 genes had no effect, consistent with their MCL1 dependence. Moreover, we confirmed that the loss of cell viability in OPM-2 cells is mediated by apoptosis, as co-incubating the cells with a broad-spectrum caspase inhibitor (Q-VD-OPh) ameliorated cell death (Figure 3B). We obtained similar results with another MCL1-dependent cell line, H929 (data not shown).
Our studies with small molecule inhibitors, as well as observations by others,11,12,27 had identified the KMS-12-PE cell line as one highly sensitive to BCL2 inhibition. Consistent with these observations, this cell line rapidly died when BCL2 was genetically targeted, using CRISPR/Cas9 (Figure 3C). Interestingly, this cell line was also killed when MCL1 was targeted.
Unlike OPM-2, H929, and KMS-12-PE, the viability of U266B1 cells was unaffected when BCL2, BCLXL, BCLW, MCL1, or BCL2A1 were genetically targeted on their own (Figure 3D). Interestingly, although targeting MCL1 alone had no effect in this cell line, cotreating MCL1-ablated cells with the well-validated small molecules allowed us to ascertain whether combinations of them keep U266B1 cells viable. Our data strongly suggest these cells rely on MCL1 and BCLXL, as targeting the former genetically using CRISPR/Cas9 and the latter by A5463 cotreatment rapidly killed U266B1 cells (Figure 3E). However, the combined targeting of MCL1 by CRISPR/Cas9 and BCL2 with ABT-199 had no effect.
As there was no adverse effect on the viability of the U266B1 cell line when the prosurvival BCL2 genes were singly targeted, we could readily confirm the highly efficient targeting of these genes in U266B1 cells by immunoblotting for the proteins (Figure 3F) or by DNA sequencing (Figure 3G; supplemental Figure 6-14). Moreover, the efficiency and specificity of guides targeting BCL2 or BCLXL were further confirmed in other cell lines (supplemental Figure 15) or in a recent study.22
Many myeloma cell lines were killed when expression of MCL1 was targeted genetically
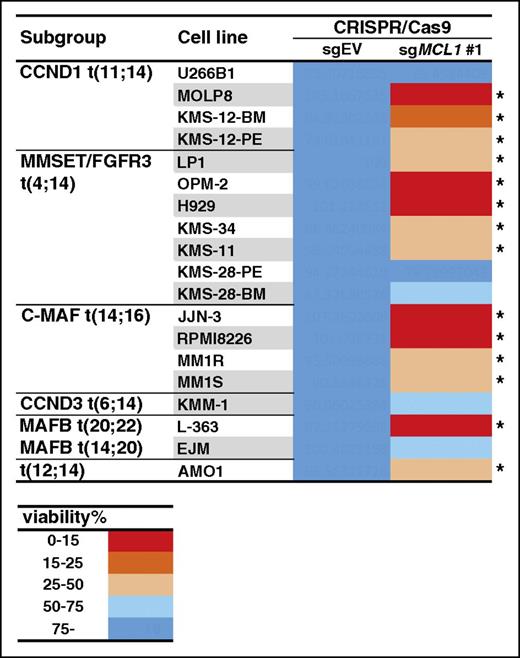
These studies (Figure 3) to assess the effect of targeting MCL1 using CRISPR/Cas9 suggested this technique has broad applicability to screen a larger panel of cell lines. First, cells that have been previously reported to rely on MCL1 (OPM-2 and H929) had significantly reduced viability when expression of MCL1 in these cells was ablated by CRSIPR/Cas9 targeting (Figure 3A-B and data not shown). Second, a cell line (KMS-12-PE) sensitive to a small molecule BCL2 inhibitor (Figure 1B) was killed when BCL2 was targeted genetically (Figure 3C). In addition to these functional studies, our sequencing analysis revealed that a significant fraction (∼50% up to 100%) of the cells in the pools harbored mutations in the target genes (Figure 3G; supplemental Figures 6-14), suggesting we can readily screen pools of cells that conditionally express a guide RNA to MCL1 to establish whether or not they rely on this prosurvival protein. Using the sgRNA for MCL1 (guide 1) introduced into the myeloma cell lines, we found that many lines, in addition to the ones previously recognized, die when MCL1 was targeted genetically (Figure 4; supplemental Table 5): the viability of 14/19 cell lines screened was reduced by greater than 50% when MCL1 was targeted using CRISPR/Cas9.
Many human myeloma cell lines are killed when MCL1 is genetically targeted. A panel of 19 myeloma cell lines was infected with a lentivirus that encodes for sgMCL1 #1 (described in Figure 3) or, as controls, the empty vector (EV). Cell viability was determined using the CellTiter-Glo assay 72 hours after the addition of DOX to induce expression of the sgRNA. A discrete heat map representation of the data is shown: red indicates very strong killing (cell viability reduced to <15%) when expression of the sgRNA is induced. In contrast, blue (>75% cell viability) indicates that the cells tolerated the genetic targeting of MCL1. *Fourteen cell lines in which expression of the sgMCL1 #1 reduced the viability by at least 50%. The results represent the mean values of 3 triplicate wells from a single experiment; the raw data are shown in supplemental Table 5.
Many human myeloma cell lines are killed when MCL1 is genetically targeted. A panel of 19 myeloma cell lines was infected with a lentivirus that encodes for sgMCL1 #1 (described in Figure 3) or, as controls, the empty vector (EV). Cell viability was determined using the CellTiter-Glo assay 72 hours after the addition of DOX to induce expression of the sgRNA. A discrete heat map representation of the data is shown: red indicates very strong killing (cell viability reduced to <15%) when expression of the sgRNA is induced. In contrast, blue (>75% cell viability) indicates that the cells tolerated the genetic targeting of MCL1. *Fourteen cell lines in which expression of the sgMCL1 #1 reduced the viability by at least 50%. The results represent the mean values of 3 triplicate wells from a single experiment; the raw data are shown in supplemental Table 5.
However, this screen had a number of potential limitations. First, we only analyzed the effect on cell viability at a few defined times (72 and 96 hours; data for the later time are not shown). Second, although the efficiency of MCL1 mutations in the U266B1 pool that express sgMCL1 #1 was high (∼90%; supplemental Figure 12), we had not determined the mutation frequency in all the cell lines screened. Moreover, the consequences of gene deletion may not be the same as using a small molecule to disrupt protein function.
Given these caveats, we adopted an alternative approach to validate our findings obtained using CRISPR/Cas9 genome editing (Figure 4), with the goal of definitively establishing whether or not MCL1 has a central role in maintaining the survival of these myeloma cell lines.
An orthogonal approach to target the prosurvival BCL2 proteins with peptidyl antagonists
To target the prosurvival BCL2 proteins with an orthogonal approach that closely recapitulates the pharmacologic action of the BH3 mimetics, we exploited a recently developed panel of BIM variants that has been fully characterized (Figure 5A).21,40,41 By comparing the viability of cells inducibly expressing WT BIM or 1 of its variants, we could deduce whether the cells depend on 1 or more of the prosurvival BCL2 proteins for their survival. Another BIM variant, BIM4E, which does not bind to any of the prosurvival proteins, served as the negative control in these studies.
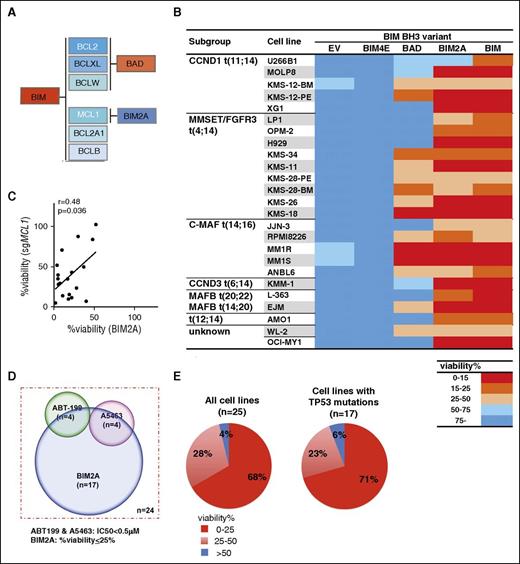
An orthogonal approach to target MCL1 confirms that most myeloma cell lines are rapidly killed when MCL1 is targeted. (A) Selectivity of wild-type BIM or its variants for the prosurvival BCL2 proteins. The BH3-only protein BIM binds to all the prosurvival BCL2 proteins with high affinity.40 BAD binds selectively to BCL2, BCLXL, and BCLW,40 whereas BIM2A, in which 2 of the key residues in the BIM BH3 domain are replaced with alanines, retains high binding affinity only for MCL1.41 BIM4E is an inert variant that fails to bind any of the prosurvival proteins.40 (B) Table summarizing the sensitivity of 25 human myeloma cell lines to expression of wild-type (WT) BIM or its variants. The myeloma cell lines were infected with lentiviruses expressing different BIM variants (A). The viability of the infected cells was determined using the CellTiter-Glo assay 24 hours after addition of DOX to induce expression of WT BIM or its variants, and normalized to that observed when the inert BIM variant (BIM4E) was expressed. A discrete heat map representation of the data are shown: red represents very strong killing (<15% viable cells), whereas blue (>75% viable cells) indicates the cells tolerated expression of the BIM variants. Notably, 17/25 lines (68% of those tested) are sensitive (viability reduced to <25%) to selective MCL1 inhibition with BIM2A. The data represent the mean values of 3 triplicate wells from a single experiment; the raw data are shown in supplemental Table 6. (C) Correlation between the sensitivity of myeloma cell lines to killing when MCL1 is genetically targeted by CRISPR/Cas9 (Figure 4) or by introduction of a MCL1-selective peptidyl antagonist, BIM2A (Figure 5B). The loss of viability of 19 myeloma cell lines after MCL1 is targeted by a sgRNA (y-axis) is correlated with the killing observed with a selective MCL1 ligand BIM2A (x-axis) (r=0.48, P = .036; the r and P values were determined with Pearson correlation analysis). (D) Most myeloma cell lines are sensitive to MCL1 inhibition. Venn diagram summarizing the sensitivity of a panel of 24 myeloma cell lines to inhibition of MCL1 (viability ≤ 25%; Figure 5B) with the selective peptidyl antagonist BIM2A, BCL2 (by ABT-199; Figure 1B) or BCLXL (by A5463; Figure 1B). Note that the WL-2 cell line, which is highly sensitive to both ABT-199 and A5463 (Figure 1B), but not to BIM2A (Figure 5B), cannot be accurately depicted, and therefore is excluded on this Venn diagram. (E) Myeloma cells lines that have deregulated TP53 function remain highly sensitive to MCL1 inhibition. Pie charts depict the degree of killing of the myeloma cell lines (left: 25 cell lines in total; right: 17 lines with mutated TP53) by the MCL1-selective peptidyl ligand BIM2A. Note that the fraction of cell lines harboring mutant TP53 that were killed by BIM2A (right) is indistinguishable from the cell line panel as a whole (left).
An orthogonal approach to target MCL1 confirms that most myeloma cell lines are rapidly killed when MCL1 is targeted. (A) Selectivity of wild-type BIM or its variants for the prosurvival BCL2 proteins. The BH3-only protein BIM binds to all the prosurvival BCL2 proteins with high affinity.40 BAD binds selectively to BCL2, BCLXL, and BCLW,40 whereas BIM2A, in which 2 of the key residues in the BIM BH3 domain are replaced with alanines, retains high binding affinity only for MCL1.41 BIM4E is an inert variant that fails to bind any of the prosurvival proteins.40 (B) Table summarizing the sensitivity of 25 human myeloma cell lines to expression of wild-type (WT) BIM or its variants. The myeloma cell lines were infected with lentiviruses expressing different BIM variants (A). The viability of the infected cells was determined using the CellTiter-Glo assay 24 hours after addition of DOX to induce expression of WT BIM or its variants, and normalized to that observed when the inert BIM variant (BIM4E) was expressed. A discrete heat map representation of the data are shown: red represents very strong killing (<15% viable cells), whereas blue (>75% viable cells) indicates the cells tolerated expression of the BIM variants. Notably, 17/25 lines (68% of those tested) are sensitive (viability reduced to <25%) to selective MCL1 inhibition with BIM2A. The data represent the mean values of 3 triplicate wells from a single experiment; the raw data are shown in supplemental Table 6. (C) Correlation between the sensitivity of myeloma cell lines to killing when MCL1 is genetically targeted by CRISPR/Cas9 (Figure 4) or by introduction of a MCL1-selective peptidyl antagonist, BIM2A (Figure 5B). The loss of viability of 19 myeloma cell lines after MCL1 is targeted by a sgRNA (y-axis) is correlated with the killing observed with a selective MCL1 ligand BIM2A (x-axis) (r=0.48, P = .036; the r and P values were determined with Pearson correlation analysis). (D) Most myeloma cell lines are sensitive to MCL1 inhibition. Venn diagram summarizing the sensitivity of a panel of 24 myeloma cell lines to inhibition of MCL1 (viability ≤ 25%; Figure 5B) with the selective peptidyl antagonist BIM2A, BCL2 (by ABT-199; Figure 1B) or BCLXL (by A5463; Figure 1B). Note that the WL-2 cell line, which is highly sensitive to both ABT-199 and A5463 (Figure 1B), but not to BIM2A (Figure 5B), cannot be accurately depicted, and therefore is excluded on this Venn diagram. (E) Myeloma cells lines that have deregulated TP53 function remain highly sensitive to MCL1 inhibition. Pie charts depict the degree of killing of the myeloma cell lines (left: 25 cell lines in total; right: 17 lines with mutated TP53) by the MCL1-selective peptidyl ligand BIM2A. Note that the fraction of cell lines harboring mutant TP53 that were killed by BIM2A (right) is indistinguishable from the cell line panel as a whole (left).
As anticipated, all the cell lines were killed by BIM expression, suggesting they rely on 1 or more of the prosurvival BCL2 proteins for their viability (Figure 5B). We next focused on the subset (6/25 cell lines) that was sensitive to BIMBAD (arbitrarily defined as percentage viability ≤ 25%) and compared them with the activity of ABT-199, which targets BCL2, or A5463, which targets BCLXL. About half the cells lines sensitive to BIMBAD were sensitive to ABT-199, whereas the other half was killed by A5463 (supplemental Figure 16). This is anticipated, as BIMBAD binds both BCL2 and BCLXL.40 Given these encouraging results, we exploited the MCL1-selective activity of BIM2A in the follow-up studies.
Most myeloma cell lines are rapidly killed when MCL1 is targeted by a selective peptidyl ligand, BIM2A
Very interestingly, we found that expression of the MCL1 selective ligand BIM2A rapidly killed a sizeable fraction (17/25 cell lines) of the myeloma cell line panel (Figure 5B; supplemental Table 6). Moreover, the activity observed with the peptidyl antagonist BIM2A correlated with the genetic targeting of MCL1 by CRISPR/Cas9 (Figure 5C), whereas the killing by BIMBAD, which does not target MCL1, did not (supplemental Figure 17).
From these approaches, using small molecule inhibitors (Figure 1), gene editing (Figures 3-4), or a peptide-based approach (Figure 5), we discerned which prosurvival protein is critical for the myeloma cell lines. Although there is some overlap (Figure 5D), it appears that most myeloma cell lines are readily killed when MCL1 is targeted (Figure 5B), whereas BCL2 or BCLXL play lesser roles (Figures 1C and 5B). Using BIM2A, strong killing (defined as viability of ≤25% of the control) was observed in 17/25 (∼68%) cell lines, and moderate killing (viability ≤50%) was observed in another 7/25 (∼28%) cell lines. We conclude that a highly significant fraction of the myeloma cell lines will be susceptible to apoptosis when MCL1 is targeted. Importantly, even cell lines that harbor the poor prognostic t(4;14) chromosomal translocation were readily killed by BIM2A (Figure 5B), and the cell lines were killed regardless of their TP53 status (Figure 5E).
Targeting MCL1 in vivo
Given the key role of MCL1 in myeloma, according to our results (Figures 3-5), and as in vitro studies cannot accurately reflect the in vivo scenario in which microenvironmental factors can modulate therapeutic responses,42,43 we next asked whether targeting this prosurvival protein might ameliorate the disease in vivo. To undertake these studies, we focused on 2 myeloma cell lines which we, and others, have identified to be reliant on MCL1: AMO1 (Figures 4-5) and H929 (Figures 2-5, and data not shown).33,35 We exploited our panel of BIM variants (Figure 5A), as the available small molecule inhibitors are not suitable (Figure 2) for in vivo studies.
Strikingly, expression of BIM2A to inhibit MCL1 in AMO1 (Figure 6; supplemental Figure 18) significantly delayed disease progression compared with mice expressing the inert BIM variant, or when BIM2A was not expressed. In the mice inoculated with the AMO1 cell line, we found significantly reduced overall disease burden (Figure 6A-B), CD38+ plasma cells in the peripheral circulation, bone marrow involvement, or serum paraprotein levels (Figure 6C-D). Equally impressive results were observed in mice inoculated with another cell line, H929 (supplemental Figure 19).
Inhibiting MCL1 suppresses the growth of the myeloma cell line AMO1 in vivo. (A) Expression of the MCL1-selective ligand BIM2A slows the growth of AMO1 myeloma cells in vivo. Here, 5 × 106 AMO1 cells engineered to coexpress the luciferase gene and the BIM variants were injected intravenously into female NSG mice. Ten days after tumor inoculation (indicated by an arrow, ↑), the mice were divided into those fed with DOX-containing (+: to induce expression of the BIM variant) or normal (−) food, and the mice then were imaged every 3 days afterward on days 3, 6, and 9. Compared with the mice expressing BIM4E or the control mice fed with normal food, significant suppression of tumor growth was observed in mice expressing BIM2A. The results are representative of 2 independent experiments (n = 3 mice in each group). The average luciferase activity was calculated from the dorsal, lateral, and ventral views of the mice. Detailed results are shown in supplemental Figure 18. (B) Representative of images of the mice described in A 9 days after commencing DOX treatment. (C) Expression of BIM2A slows disease progression in vivo. The infiltration of myeloma cells (CD38+) in the peripheral blood (upper), bone marrow (middle), or serum paraprotein (IgAκ) levels (bottom) were determined 0 to 10 days after the mice were commenced on treatment with DOX (n = 3 mice in each group). Note the delayed disease progression when expression of BIM2A was induced. (D) BIM2A ameliorates the infiltration of AMO1 myeloma cells into the bone marrow. Representative histological images (H&E staining; ×0.4 or ×5 magnification) for detection of bone marrow infiltration with myeloma cells (pale areas) taken from the legs of representative mice. The regions marked with an arrow (left) were enlarged for higher magnification (right; ×5 instead of ×0.4).
Inhibiting MCL1 suppresses the growth of the myeloma cell line AMO1 in vivo. (A) Expression of the MCL1-selective ligand BIM2A slows the growth of AMO1 myeloma cells in vivo. Here, 5 × 106 AMO1 cells engineered to coexpress the luciferase gene and the BIM variants were injected intravenously into female NSG mice. Ten days after tumor inoculation (indicated by an arrow, ↑), the mice were divided into those fed with DOX-containing (+: to induce expression of the BIM variant) or normal (−) food, and the mice then were imaged every 3 days afterward on days 3, 6, and 9. Compared with the mice expressing BIM4E or the control mice fed with normal food, significant suppression of tumor growth was observed in mice expressing BIM2A. The results are representative of 2 independent experiments (n = 3 mice in each group). The average luciferase activity was calculated from the dorsal, lateral, and ventral views of the mice. Detailed results are shown in supplemental Figure 18. (B) Representative of images of the mice described in A 9 days after commencing DOX treatment. (C) Expression of BIM2A slows disease progression in vivo. The infiltration of myeloma cells (CD38+) in the peripheral blood (upper), bone marrow (middle), or serum paraprotein (IgAκ) levels (bottom) were determined 0 to 10 days after the mice were commenced on treatment with DOX (n = 3 mice in each group). Note the delayed disease progression when expression of BIM2A was induced. (D) BIM2A ameliorates the infiltration of AMO1 myeloma cells into the bone marrow. Representative histological images (H&E staining; ×0.4 or ×5 magnification) for detection of bone marrow infiltration with myeloma cells (pale areas) taken from the legs of representative mice. The regions marked with an arrow (left) were enlarged for higher magnification (right; ×5 instead of ×0.4).
Low-passage myeloma cell lines are also highly sensitive to MCL1 inhibition
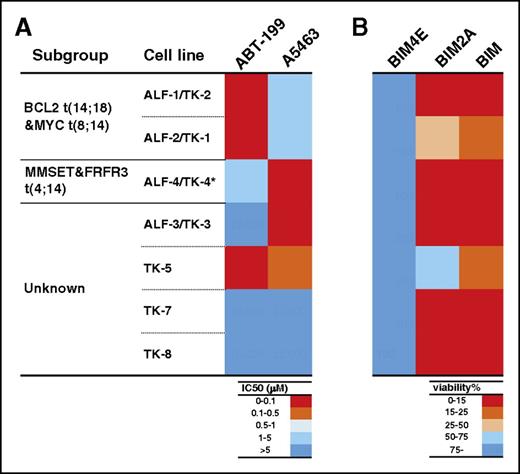
Our data from studies, both in vitro (Figures 4-5) and in vivo (Figure 6; supplemental Figures 18-19), using the panel of cell lines, strongly suggests MCL1 is an important therapeutic target to consider for myeloma. To extend our studies beyond these immortalized cell lines that are well adapted to growth under tissue culture conditions, we evaluated the effect of BCL2, BCLXL, or MCL1 inhibition on a panel of low-passage cell lines, some with complex poor-risk karyotypes.19 Three of 7 and 2/7 lines were readily killed by inhibition of BCL2 or of BCLXL, respectively (Figure 7A; supplemental Table 7). Strikingly, we found that 5/7 of these cell lines tested were readily killed (viability, ≤25%) by expressing the MCL1-selective ligand BIM2A (Figure 7B; supplemental Table 7), further defining MCL1 as a therapeutic target of high potential in multiple myeloma.
The MCL1-selective peptidyl antagonist BIM2A kills low-passage tumor cells derived from patients with multiple myeloma. The sensitivity of 7 low-passage myeloma cell lines to the selective inhibition of BCL2, BCLXL, or MCL1 was tested using either BH3 mimetics (A) or BIM variants (B). (A) Briefly, the 7 cell lines were treated with ABT-199 (to target BCL2) or A5463 (to target BCLXL), and their sensitivity to these agents was summarized in a heat map. (B) To evaluate their sensitivity to MCL1 inhibition, the lines were also infected with lentiviral vectors that inducibly express BIM4E (inert variant), BIM2A (MCL1-selective), or BIM (to target all the prosurvival proteins) and the sensitivity summarized in a heat map. Notably, 5/7 lines were killed when MCL1 was inhibited with the MCL1-selective ligand BIM2A. The data show the mean values ± 1 SD from 3 independent experiments. ALF1-1/TK-2 and ALF-2/TK-1 were derived from the peripheral blood and bone marrow from the same patient19 ; they harbor the t(14;18) translocation. *The ALF-4/TK-4 cell line harbors the del17p mutation.
The MCL1-selective peptidyl antagonist BIM2A kills low-passage tumor cells derived from patients with multiple myeloma. The sensitivity of 7 low-passage myeloma cell lines to the selective inhibition of BCL2, BCLXL, or MCL1 was tested using either BH3 mimetics (A) or BIM variants (B). (A) Briefly, the 7 cell lines were treated with ABT-199 (to target BCL2) or A5463 (to target BCLXL), and their sensitivity to these agents was summarized in a heat map. (B) To evaluate their sensitivity to MCL1 inhibition, the lines were also infected with lentiviral vectors that inducibly express BIM4E (inert variant), BIM2A (MCL1-selective), or BIM (to target all the prosurvival proteins) and the sensitivity summarized in a heat map. Notably, 5/7 lines were killed when MCL1 was inhibited with the MCL1-selective ligand BIM2A. The data show the mean values ± 1 SD from 3 independent experiments. ALF1-1/TK-2 and ALF-2/TK-1 were derived from the peripheral blood and bone marrow from the same patient19 ; they harbor the t(14;18) translocation. *The ALF-4/TK-4 cell line harbors the del17p mutation.
Discussion
The main focus of our study was to definitively establish which of the prosurvival BCL2 proteins is required to maintain the viability of multiple myeloma cells and to establish a hierarchy of targets for clinical evaluation. Using pharmacologic (Figure 1) as well as genetic (Figure 4) and peptide-based (Figure 5) approaches, we showed that the majority of the myeloma cell lines tested, including low-passage ones, rely on the prosurvival protein MCL1, with a smaller fraction dependent on BCL2 or on BCLXL (Figures 5D and 7). Taken together with recent findings,44 our results provide strong impetus for testing potent and selective MCL1 inhibitors for the treatment of this disease. Although some BH3 mimetics such as ABT-199 (venetoclax) and ABT-263 (navitoclax) are undergoing clinical trials, ones to target MCL1 are in a much earlier developmental stage.33,45,46
The ever-expanding repertoire of small molecule inhibitors of 1 or more of the prosurvival BCL2 proteins is proving invaluable to preclinical studies to establish which of BCL2 proteins a particular tumor relies on.11,17,27,44,47,48 Importantly, the potency, selectivity, and specificity of these compounds must be determined in mechanism-of-action studies; our finding that a significant subset of the myeloma cell lines is susceptible to BCLXL inhibition (Figure 1) is not widely appreciated. Our conclusion relies on using a potent inhibitor of BCLXL, A-1155463, which is significantly more active than ABT-737 (supplemental Figure 3; supplemental Table 4). Precisely why this is the case is unclear and is the subject of our ongoing studies.
Although similar studies using small molecule inhibitors to selectively target MCL1 were not feasible, we were able to exploit the power of CRISPR/Cas9 genome editing to rapidly screen the panel of myeloma cell lines (Figure 4). This technique proved rapid and reliable, as we were able to confirm our findings using a peptide ligand BIM2A that is highly selective for MCL1 (Figure 5). Thus, our studies, using 2 distinct approaches (CRISPR/Cas9 genome editing to target MCL1 or the MCL1-selective ligand BIM2A), lend support to previous studies that suggested the potential importance of MCL1 for multiple myeloma.30,31,35 We anticipate that our studies pinpointing the central role of MCL1 in multiple myeloma, including in vivo studies (Figure 6), are very likely to closely mirror the clinical setting.
Our results also draw attention to ongoing efforts attempting to identify optimal targets for developing novel cancer therapeutics. Although the notion that targeting the molecular lesion driving a cancer is compelling and validated, such as the case with targeting the BCR-ABL fusion oncoprotein in chronic myelogenous leukemia or mutant B-RAF V600E in melanoma, it has been argued that myeloma cells may be more susceptible to some classes of targeted therapeutics, such as the proteasome inhibitors or to immune modulator agents, because they retain fundamental properties of their normal cellular counterparts, the plasma cells, from which they are derived.49 In this regard, it is noteworthy that previous biological studies, mainly in the mouse, have identified MCL1 as the critical survival factor required to sustain plasma cells.28,29 As such, one could hypothesize that the targeting of critical prosurvival proteins such as MCL1 will prove to be agnostic to the recognized adverse biological features of myeloma50 that continue to define the limitations of presently available immune modulator agents and proteasome inhibitor-based therapeutic approaches.
In summary, our results highlight the pressing urgency for the development of high-quality, potent inhibitors of MCL1 for efficacy and safety studies, particularly in the context of treating multiple myeloma. MCL1 should be added to the list of highly promising targets for the development of novel therapies in this disease, and we anticipate that it would prove clinically effective, provided safety concerns can be adequately addressed.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank J. M. Adams, S. Cory, A. Strasser, M. F. van Delft, and J. G. Zhang for discussions and suggestions; R. Anderson and W. Welch for gifts of reagents; D. Cooper, C. Hay, S. Oliver, and G. Siciliano for animal husbandry; and AbbVie for providing ABT-737, ABT-199, and A-1155463.
This work is supported by scholarships, fellowships, and grants from the Australian National Health and Medical Research Council (research fellowships to A.W.R. and D.C.S.H.; project grant 1057742 [D.C.S.H.]; program grants 1016647 and 1016701; and Independent Research Institutes Infrastructure Support Scheme [grant 9000220]), the Cancer Council Victoria (grant-in-aid to A.W.R. and D.C.S.H.), the Leukemia and Lymphoma Society (Specialized Centers of Research grants 7001-13), the Australian Cancer Research Foundation, a Victorian State Government Operational Infrastructure Support grant, and the China Scholarship Council (award to Y.Y.). A.W.R. holds the Metcalf Chair of Leukaemia Research at the University of Melbourne.
Authorship
Contribution: J.-N.G., A.W.R., and D.C.S.H. devised the study; J.-N.G., T.K., D.S., Y.Y., and C.D.R. performed the experiments; J.-N.G., T.K., D.S., S.L.K., and M.J.H. developed methodology; T.K. and A.S. provided the low-passage myeloma cell lines; J.-M.G. and G.L. provided A-1331852; J.-N.G., T.K., D.S., Y.Y., C.D.R., A.S., A.W.R., and D.C.S.H. analyzed the data; J.-N.G. wrote the first draft of the manuscript, which was edited and revised by A.W.R. and D.C.S.H.; all authors contributed to review and analysis of data in the manuscript. The study was supervised by A.W.R. and D.C.S.H.
Conflict-of-interest disclosure: J.-N.G., D.S., C.D.R., S.L.K., G.L., M.J.H., A.W.R., and D.C.S.H. are employees of the Walter and Eliza Hall Institute of Medical Research, which receives research funding and milestone payments in relation to venetoclax (ABT-199). The laboratories of A.W.R. and D.C.S.H. receive research funding from Servier. The rest of the authors declare no competing financial interests.
The current affiliation for J.-M.G. is SYNthesis Med Chem, Melbourne, Australia.
Correspondence: Andrew W. Roberts, The Walter and Eliza Hall Institute of Medical Research, 1G Royal Parade, Parkville, VIC 3052, Australia; e-mail: roberts@wehi.edu.au; and David C. S. Huang, The Walter and Eliza Hall Institute of Medical Research, 1G Royal Parade, Parkville, VIC 3052, Australia; e-mail: huang_d@wehi.edu.au.
References
Author notes
A.W.R. and D.C.S.H. are joint senior authors.