Key Points
tTregs express high miR-146b levels and downregulate TRAF6 mRNA and NF-κB activation, which is essential for tTreg function.
miR-146b antagomir enhances in vitro and in vivo tTreg suppression and persistence as well as xenogenic GVHD lethality.
Abstract
CD4+CD25+FoxP3+ thymic-derived regulatory T cells (tTregs) are indispensable for maintaining immune system equilibrium. Adoptive transfer of tTregs is an effective means of suppressing graft-versus-host disease (GVHD) in murine models and in early human clinical trials. Tumor necrosis factor receptor-associated factor 6 (TRAF6), an ubiquitin-conjugating enzyme that mediates nuclear factor κB (NF-κB) activation, plays an essential role in modulating regulatory T cell survival and function. MicroRNAs (miRNAs) are noncoding RNAs, which mediate RNA silencing and posttranscriptional gene repression. By performing comprehensive TaqMan Low Density Array miRNA assays, we identified 10 miRNAs differentially regulated in human tTreg compared with control T cells. One candidate, miR-146b, is preferentially and highly expressed in human naive tTregs compared with naive CD4 T cells. miRNA prediction software revealed that TRAF6 was the one of the top 10 scored mRNAs involved tTreg function with the highest probability as a potential miR-146b target. Antagomir-mediated knockdown of miRNA-146b, but not another miRNA-146 family member (miRNA-146a), enhanced TRAF6 expression. TRAF6, in turn, increases NF-κB activation, which is essential for tTreg function as well as Foxp3 protein and antiapoptotic gene expression, and downregulates proapoptotic gene expression. miR-146b knockdown increased the nuclear localization and expression of genes regulated by NF-κB, which was associated with enhanced tTreg survival, proliferation, and suppressive function measured in vitro and in vivo. TRAF6 inhibition had the opposite effects. We conclude that an miR-146b–TRAF6-NF-κB–FoxP3 signaling pathway restrains regulatory T cell survival, proliferation, and suppressor function. In vitro exposure of human tTregs to miR-146b antagomirs can be exploited to improve the clinical efficacy of human adoptive tTreg transfer in a GVHD setting.
Introduction
Graft-versus-host disease (GVHD) is a multi–organ system complication of allogeneic hematopoietic stem cell transplantation that is due to unperturbed donor anti–host T cell destructive responses.1-3 Immunosuppressive drugs are used to prevent GVHD but have been neither uniformly successful nor free of significant side effects. CD4+CD25+ thymic-derived regulatory T cells (tTregs), which express the transcription factor FoxP3 (positive) and are CD127 low, control immune homeostasis. In rodents, donor graft tTreg supplementation has been shown to be highly effective in suppressing GVHD lethality.4-6 In the clinic, the infusion of tTregs can reduce the incidence and severity of acute GVHD.6-10 High ratios of donor tTregs to conventional T cells were needed for maximal GVHD protection. Thus, an approach to imbue tTregs with stronger suppressive function and proliferative ability would be beneficial for preventing GVHD as well as preventing solid organ graft rejection or treating autoimmune disease.11,12
MicroRNAs (miRNAs), a family of ∼22-nt noncoding RNAs, regulate gene expression by binding to messenger RNA (mRNA) though nucleotides at the 5′ end of the miRNA. They operate on RNA silencing and posttranscriptional gene repression by matching with the seed region, mostly situated at positions 2 to 7 from the miRNA 5′ end, and are essential for binding of the miRNA to the mRNA.13,14 Multiple mechanisms have been implicated in tTreg development and suppressive function, including more recently miRNA’s effects on posttranscriptional gene regulation.15-18 The miR-146 family includes 2 main members, miR-146a and miR-146b, which are both highly expressed and play important roles in cell proliferation and function in diverse cell types, including T and B cells. Studies of breast cancer cells have shown a Foxp3-dependant increase in the expression of miR-146b and, to a lesser extent, miR-146a,19 which results in reduced breast cancer cell proliferation and enhanced apoptosis via negative regulation of nuclear factor κB (NF-κB). Even though they share the same seed regions, posttranscriptional processing mechanisms and consequently functions may be distinct due to different genomic locations (chromosomes 5 and 10, respectively). Whereas activation of NF-κB negatively correlates with miR-146a expression, miR-146b-5p is induced by cytokines that activate signal transducer and activator of transcription 3 (STAT30 (interleukin-6 [IL-6]) or STAT1 (interferon-γ [IFN-γ]).20-23 Lipopolysaccharide induction of miR-146a was observed in bone marrow–derived macrophages, but IL-10 release from macrophages leads to expression of miR-146b, but not miR-146a, suggesting different signaling pathways for miR-146a and miR-146b.24 Knockout of miR-146a results in excessive regulatory T cell (Treg) IFN-γ production and reduced Treg-mediated Th1 response inhibition, resulting in fatal autoimmune disease.25 Furthermore, CD4+ T cells were moderately reduced in miR-146b, but not miR-146a, transgenic mice due to impaired ability to expand in response to T cell receptor (TCR) stimulation.26 TRAF6 plays an essential role in NF-κB activation and Treg proliferation and function. TRAF6 deficiency results in immune tolerance imbalance and autoimmune disease.27,28 Although TRAF6 is a predicted target of miR-146a/b in human and murine tumor cells29,30 and Tregs in mice,25 no significant correlation has been observed between miR-146a and TRAF6 in human T cells.31 Thus, identifying the roles of miRNA-146b–regulated pathways in tTregs, which might be distinct from miR-146a, could further be used to enhance Treg suppressive function and/or stability and improve the therapeutic potential of Treg adoptive cell therapy.
By performing comprehensive TaqMan Low Density Array miRNA assays, we identified 10 miRNAs differentially regulated in human tTregs compared with control T cells that might play important roles in Treg stability and function. We demonstrate that knockdown of miR-146b increases TRAF6 expression in human tTregs, leading to NF-κB activation and enhanced FoxP3 expression, suppressive function, and proliferative ability in vitro and in vivo. Together, these data provide both an insight into human tTreg function and a new approach in the clinic to improve the efficacy of tTregs in preventing GVHD.
Materials and methods
More information regarding mice, cell purification and culture, flow cytometry, Imagestream analysis, gene expression analysis, the miRNA target prediction and reporter assay, the suppression assay, xenogenic GVHD studies, and statistics can be found in supplemental Materials and methods (available on the Blood Web site).
Treatment of tTregs and CD4+ T cells
tTregs and CD4+ T-cell cultures, generated as described in supplemental Methods, were frozen on days 14 and 7, respectively. Frozen cells were thawed and restimulated with anti-CD3/CD28 monoclonal antibody–coated Dynabeads (Thermo Fisher Scientific, Carlsbad, CA) at 1:3 (cell-to-bead) ratios in the presence of recombinant IL-2. After 6 days, cultures were washed and resuspended at 1 × 106 cells/mL, and nanoparticle-encapsulated RNA (50 nM scramble/antagomir; EXIQON, Woburn, MA; supplemental Table 2), TRAF6 inhibitor (8 µM; EMD Millipore, Darmstadt, Germany), or NF-κB inhibitor (3 µM or 6 µM PS-1145; Millennium Pharmaceuticals, Cambridge, MA) was added. Dimethylsulfoxide as the vehicle used as a control. Cells were cultured for another 2 days without further manipulation and harvested and assayed as listed. For some experiments, tTregs were kept in culture longer (as indicated) without further antagomir addition.
Results
Identification of human tTreg-specific miRNA
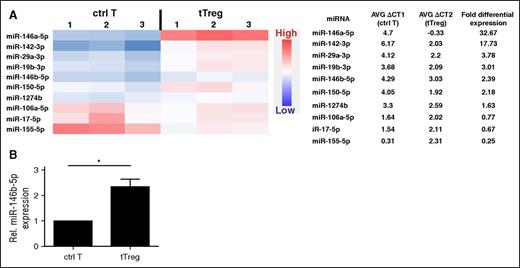
Changes in miRNA expression are observed during T-cell differentiation.32 To identify potential human tTreg-specific miRNA, we compared miRNA expression between in vitro–expanded naive CD4 T cells (CD4+25−127+45RA+; control T cells) and naive tTregs (CD4+25++127−45RA+). TaqMan Low Density Array was used to reveal differential miRNA expression between human naive tTregs and CD4 T cells. RNU44, an ubiquitously expressed small nucleolar RNA, was used to normalize expression between the cell types. After analyzing 768 miRNAs by ranking fold expression, the top 10 differential miRNAs between tTregs and CD4 T cells were chosen for further analysis (Figure 1A). Two miRNAs, which were preferentially expressed (>10.0-fold) in tTregs (miR-146a-5p and miR-142-3p), have been implicated in tTreg development and/or function. miR-146a-5p has proved to be essential for Treg development, phenotype, and function, while miR-142-3p negatively regulates tTreg suppressive function by restricting the AC9/cyclic adenosine 5′-monophosphate pathway.25,33-35 Four miRNAs (miR-29a-3p, miR-19b-3p, miR-150-5p, and miR-146b-5p), which had differential expression >2.0-fold, also have known functions in tTregs; miR-29a and miR-150 promote Treg differentiation, while miR-19b negatively regulates Treg induction from naive T cells.18,36-39
miRNA profiling of expanded naïve CD4+ T cells and naive tTregs demonstrates stronger expression of miR-146b in human tTregs (n = 3). Naive T cells (CD4+25-127+45RA+) and naive tTregs (CD4+25++127-45RA+) were sort-purified and expanded in vitro. miRNA expression in control T cells (ctrl T) and tTregs was determined by miRNA TaqMan Low Density Array. (A) After analyzing 768 miRNAs, the top 10 differential miRNAs between tTregs and control T cells were gated for further analysis by heatmap (left) and average (AVG) fold differential expression (right) (P < .05) . (B) Relative (Rel.) differential expression of miR-146b in control T cells and tTregs (n = 3) was confirmed by RT-PCR *P < .05.
miRNA profiling of expanded naïve CD4+ T cells and naive tTregs demonstrates stronger expression of miR-146b in human tTregs (n = 3). Naive T cells (CD4+25-127+45RA+) and naive tTregs (CD4+25++127-45RA+) were sort-purified and expanded in vitro. miRNA expression in control T cells (ctrl T) and tTregs was determined by miRNA TaqMan Low Density Array. (A) After analyzing 768 miRNAs, the top 10 differential miRNAs between tTregs and control T cells were gated for further analysis by heatmap (left) and average (AVG) fold differential expression (right) (P < .05) . (B) Relative (Rel.) differential expression of miR-146b in control T cells and tTregs (n = 3) was confirmed by RT-PCR *P < .05.
miR-146b-5p drew our attention for deeper investigation. Although some reports found miR-146a-5p positively regulates tTreg function in mice,25 others found miR-146a and/or 146b knockdown had no impact on human Treg function.40 Therefore, we hypothesized that miR-146b-5p and miR-146a-5p might have opposite roles in controlling tTreg function. To confirm the higher expression in tTregs, differential expression of miR-146b-5p was validated by quantitative reverse transcription polymerase chain reaction (RT-PCR) (Figure 1B) and hence chosen for this study.
Knockdown of miR-146b-5p enhances FoxP3 expression, viability, expansion and suppressive function in vitro
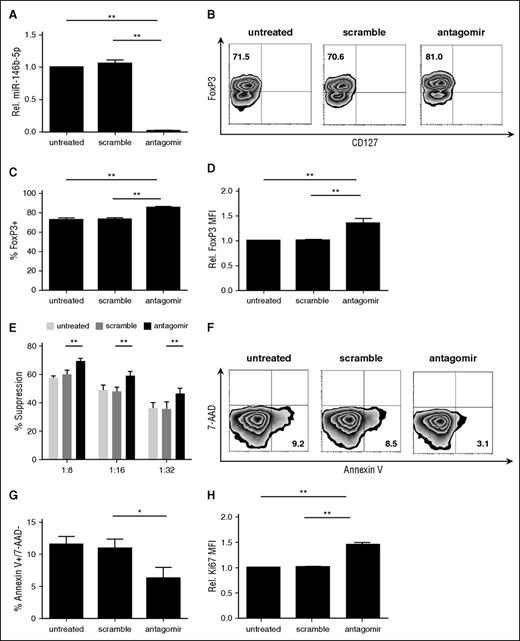
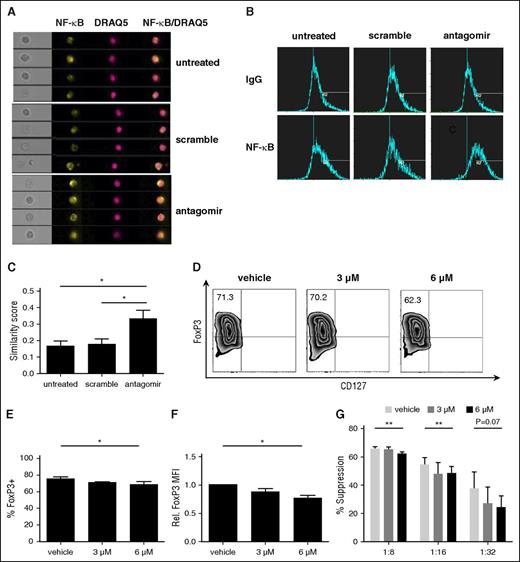
To determine whether miR-146b-5p affected human tTreg phenotype or suppressive function, tTregs were incubated with nanoparticle-encapsulated miR-146b-5p antagomir as described in supplemental materials and Methods. Scrambled miRNA was used as a control. miR-146b-5p knockdown efficiency was ≥95% (Figure 2A) as assessed by quantitative RT-PCR. Knockdown of miR-146b-5p in tTregs increased Foxp3 protein expression at the population (85.5% ± 3.5% vs 73.1% ± 4.6%, P < .05; Figure 2B-C) and per-cell (Figure 2D) levels compared with the scrambled group. We also compared the in vitro suppressive function of untreated tTregs with those treated with scrambled or miR-146b-5p antagomir. Using a carboxyfluorescein diacetate succinimidyl ester (CFSE)-based proliferation assay with tTreg to peripheral blood mononuclear cell (PBMC) ratios ranging from 1:8 to 1:32, antagomir treatment was found to significantly enhance tTreg suppressive function at each tTreg:PBMC ratio tested (Figure 2E), with an approximately twofold increase in efficacy (ie, antagomir-treated tTregs at 1:16 were as suppressive as scramble-treated tTregs at 1:8). One previous study suggested that CD4 T cells from miR-146b (but not miR-146a) transgenic mice were significantly impaired in their ability to expand in response to in vitro TCR stimulation due to a combination of increased apoptosis and a defective entrance into S phase.26 Therefore, we tested whether miR-146b-5p antagomir treatment would have an impact on in vitro tTreg expansion. Although no significant difference in expansion was observed during the initial 2-day treatment period, antagomir-treated tTreg cultures showed increased viability and staining for Ki-67, a marker of proliferation (Figure 2F-H). Cultures were continued without further antagomir treatment to assess long-term cell accumulation. By day 8 posttreatment, antagomir-treated tTregs had expanded significantly more than untreated tTregs or those treated with scrambled RNA (supplemental Figure 1A). Effects of antagomir treatment were durable, as miR-146b expression was still reduced 70% on day 8 (not shown) and treated tTregs maintained their higher Foxp3 expression and suppressive function (supplemental Figure 1B-E). Therefore, knockdown of miR-146b-5p favors FoxP3 expression, expansion, and suppressive function and attenuates apoptosis protein.
tTregs treated with miR-146b-5p antagomir show increased FoxP3 and Ki67 expression and viability with enhanced suppressive function. Naive peripheral blood tTregs were sort-purified, expanded in vitro, and treated with or without scramble/antagomir for the final 2 days of culture. (A) miRNA was purified from each culture, and miR-146b expression was assessed by RT-PCR to determine knockdown efficiency (n = 3). (B) Representative example of Foxp3 vs CD127 staining in tTregs treated with antagomir compared with untreated and scramble groups (gated on CD4+ cells). Summary of overall percentage of Foxp3+CD127− cells (C) and level of Foxp3 expression (D) in tTregs from each group (n = 5). (E) Percent suppression of in vitro, anti-CD3–mediated CD8+ T cell proliferation at ratios from 1:8 to 1:32 (tTregs:PBMCs) as determined by CFSE dye dilution (n = 5). (F, G) Representative flow images (F) and statistical analysis (G) of apoptosis tests were performed to analyze the survival ability in vitro. Viability was significantly enhanced after antagomir treatment (n = 3). (H) A higher relative Ki67 mean fluorescence intensity level was found after antagomir treatment (n = 5). Values indicate mean ± standard error of the mean (SEM) of these experiments. *P < .05; **P < .01.
tTregs treated with miR-146b-5p antagomir show increased FoxP3 and Ki67 expression and viability with enhanced suppressive function. Naive peripheral blood tTregs were sort-purified, expanded in vitro, and treated with or without scramble/antagomir for the final 2 days of culture. (A) miRNA was purified from each culture, and miR-146b expression was assessed by RT-PCR to determine knockdown efficiency (n = 3). (B) Representative example of Foxp3 vs CD127 staining in tTregs treated with antagomir compared with untreated and scramble groups (gated on CD4+ cells). Summary of overall percentage of Foxp3+CD127− cells (C) and level of Foxp3 expression (D) in tTregs from each group (n = 5). (E) Percent suppression of in vitro, anti-CD3–mediated CD8+ T cell proliferation at ratios from 1:8 to 1:32 (tTregs:PBMCs) as determined by CFSE dye dilution (n = 5). (F, G) Representative flow images (F) and statistical analysis (G) of apoptosis tests were performed to analyze the survival ability in vitro. Viability was significantly enhanced after antagomir treatment (n = 3). (H) A higher relative Ki67 mean fluorescence intensity level was found after antagomir treatment (n = 5). Values indicate mean ± standard error of the mean (SEM) of these experiments. *P < .05; **P < .01.
Previous murine studies suggested that miR-146a-5p was involved in Th1 responses and its deficiency led to dysregulated IFN-γ production.25,41 However, few expanded human tTregs expressed IFN-γ under our conditions, and expression was not affected by miR-146b-5p antagomir treatment (supplemental Figure 2). Because control CD4 T cells also expressed miR-146b-5p, albeit at low levels, it seemed possible that downmodulation of this miRNA would induce Foxp3 expression and suppressive function. Naive CD4 T cells were purified, expanded in vitro using conditions similar to those in the original array study, and treated with miR-146-5p antagomir for the final 2 days. miR-146b antagomir treatment had no effect on FoxP3 expression or suppressive function in control CD4 T cells (supplemental Figure 3). We hypothesize that the lack of an effect of miR-146b antagomir on conventional T cells is likely due to low miR-146b expression, perhaps related to a lack of Foxp3-mediated miR-146b expression. Alternatively, this difference could be due to the fact that TCR signaling, including TRAF6 and NF-κB activation, is known to be differentially controlled in conventional T cells compared with tTreg. In supplemental Figure 3, no increase in suppressive function was observed. However, a low level of suppression was observed in all effector T cell cultures. This most likely due to competition for the stimulatory anti-CD3 beads, especially because the in vitro–expanded effector T cell cells are already in cycle and are physically larger (and thus express more TCR) than the T cells present in the directly ex vivo PBMC cultures. These data indicate that knockdown of miR-146b-5p in human tTregs, but not control CD4 T cells, increases FoxP3 expression and suppressive function in vitro without increasing IFN-γ production.
TRAF6 is a direct target of miR-146b-5p, and knockdown of miR-146b-5p increases TRAF6 expression in human tTregs
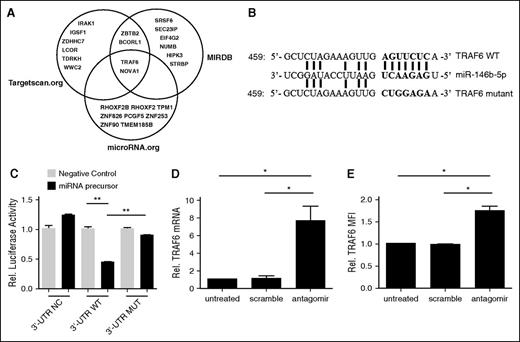
To identify potential miR-146b targets, we used miRNA prediction software (targetscan.org, MIRDB.org, and microRNA.org) to reveal the potential targeted miRNAs. After sorting top 10 predicted miRNAs in each software package, we found that TRAF6 was the one mRNA involved tTreg function with highest possibility (Figure 3A). All 3 sets of software identified potential miR-146b-5p binding sites (7 nt) in the 3′ UTR of TRAF6 (Figure 3B). Consistent with this hypothesis, TRAF6 expression was decreased in miR-146b transgenic mice,26 and inversely correlated with miR-146b-5p expression in umbilical vein endothelial cells (HUVECs) and dendritic cells (DCs).42,43 To confirm the predicted miR-146b-5p binding site in TRAF6, HEK293 cells were transiently transfected with pGL3 firefly luciferase (ff-luc)reporter plasmids with no insert, or with the wild-type (WT) or mutated (MUT) 3′ UTR sequences of TRAF6 (supplemental Table 1), along with 25 nM miR-146b-5p or negative control RNA. As shown in Figure 3C, coincubation of control RNA or miR-146b did not show any significant effects (P = .07). While coincubation of control RNA with WT 3′ UTR–tagged ff-luc had no effect, coincubation with miR-146b resulted in a 60% decrease in ff-luc activity (Figure 3C, middle panels). Furthermore, the specificity of this interaction was confirmed because miR-146b did not significantly decrease ff-luc activity in HEK293 cells expressing the mutant 3′ UTR (Figure 3C, right panels). While miR-146a and –miR-146b are separate gene products, they share a homologous seed region. Since miR-146a can also target TRAF6, we analyzed miR-146a expression following miR-146b antagomir treatment by quantitative RT-PCR to assess knockdown specificity. Our data demonstrate that miR-146a expression in tTregs treated with miR-146b-5p antagomir was similar to that of tTregs treated with scramble RNA (supplemental Figure 4).
TRAF6 is a direct target of miR-146b-5p and knock-down of miR-146b-5p increased TRAF6 expression in human tTregs (n = 3). To assess whether human miR-146b-5p targets TRAF6, HEK293 cells were transduced with plasmids carrying wild-type (WT) or mutant (MUT) 3′ UTR sequences from TRAF6 linked to a luciferase reporter gene. Cells were also transfected with a Renilla luciferase reporter construct for normalization. (A) Three software packages (targetscan.org, MIRDB, and microRNA.org) were used to predict the potential target mRNAs of miR-146b-5p; TRAF6 was involved in tTreg function with highest probability. (B) Schematic representation of the miR-146b-5p target sequence within the 3′ UTR of TRAF6. Two nucleotides (complementary to nucleotides 6 and 8 of miR-146b-5p) were mutated in the 3′ UTR of TRAF6. The numbers indicate the positions of the nucleotides in the reference WT sequences. (C) Activity of the luciferase gene linked to the WT or MUT 3′ UTR of TRAF6. Luciferase activity was measured after 48 hr. The mean of the results from the cells transfected by control vector was set as 100%. The data are mean and standard deviation (SD) of separate transfections (n = 3). Naive peripheral blood tTregs were sort-purified, expanded in vitro, and treated with or without miR-146b antagomir or scrambled RNA as previously described. After treatment, cultured cells were assessed for TRAF6 mRNA and protein expression by RT-PCR or flow cytometry (D and E, respectively). Values indicate mean ± SEM of these experiments. *P < .05; **P < .01.
TRAF6 is a direct target of miR-146b-5p and knock-down of miR-146b-5p increased TRAF6 expression in human tTregs (n = 3). To assess whether human miR-146b-5p targets TRAF6, HEK293 cells were transduced with plasmids carrying wild-type (WT) or mutant (MUT) 3′ UTR sequences from TRAF6 linked to a luciferase reporter gene. Cells were also transfected with a Renilla luciferase reporter construct for normalization. (A) Three software packages (targetscan.org, MIRDB, and microRNA.org) were used to predict the potential target mRNAs of miR-146b-5p; TRAF6 was involved in tTreg function with highest probability. (B) Schematic representation of the miR-146b-5p target sequence within the 3′ UTR of TRAF6. Two nucleotides (complementary to nucleotides 6 and 8 of miR-146b-5p) were mutated in the 3′ UTR of TRAF6. The numbers indicate the positions of the nucleotides in the reference WT sequences. (C) Activity of the luciferase gene linked to the WT or MUT 3′ UTR of TRAF6. Luciferase activity was measured after 48 hr. The mean of the results from the cells transfected by control vector was set as 100%. The data are mean and standard deviation (SD) of separate transfections (n = 3). Naive peripheral blood tTregs were sort-purified, expanded in vitro, and treated with or without miR-146b antagomir or scrambled RNA as previously described. After treatment, cultured cells were assessed for TRAF6 mRNA and protein expression by RT-PCR or flow cytometry (D and E, respectively). Values indicate mean ± SEM of these experiments. *P < .05; **P < .01.
We next asked whether miR-146b-5p antagomir treatment affected TRAF6 expression in expanded tTregs. Figure 3D shows that although no effect was observed in the scramble group, tTregs treated with miR-146b-5p antagomir had significantly higher TRAF6 mRNA expression. In accordance with mRNA level, antagomir treatment significantly enhanced TRAF6 protein expression in tTregs compared with the untreated or scramble-treated groups (Figure 3E). Taken together, these results show that TRAF6 is a direct target of miR-146b-5p in human tTregs.
Inhibition of TRAF6 signaling impairs human tTreg expansion and Foxp3 expression and suppressive function
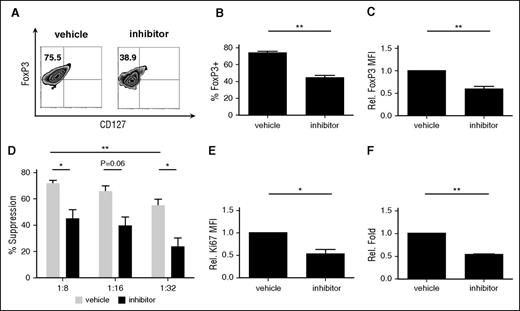
Conditional knockout of TRAF6 in murine Treg resulted in decreased Foxp3 stability and loss of in vivo suppressive function.27 We sought to determine if TRAF6 plays a similar role in human tTreg. In vitro expanded tTreg were treated with a TRAF6 signaling inhibitor (6877002)44 for the final 2 days of culture. Similar to reports for murine tTregs, inhibition of TRAF6 signaling in human tTreg cultures decreased Foxp3 protein expression both at a population level (68.0% ± 6.6% vs 46.8% ± 6.8%, P < .05, Figure 4A-B) and on an individual-cell basis (Figure 4C) compared with the vehicle group. Suppressive function was markedly downregulated after inhibitor treatment (Figure 4D). TRAF6 inhibitor also decreased proliferative ability (as assessed by Ki-67 staining) after 2 days of treatment and significantly decreased cell number after 8 days of treatment (Figure 4E-F).
Inhibition of TRAF6 signaling impairs human tTreg expansion, Foxp3 expression and suppressive function. tTregs were treated with TRAF6 inhibitor for 2 days (n = 5). (A) Representative flow figures of FoxP3+ tTregs in different groups. Foxp3 expression was significantly decreased after inhibitor treatment. (B) FoxP3+ population and (C) FoxP3 expression was measured in these groups, and the inhibitor-treated group was significantly decreased. (D) CFSE assay was performed to measure suppressive ability and showed decreased suppressive function at 1:8 and 1:32. Values indicate mean ± SEM of these experiments. (E) Ki67 expression was measured in these groups, and Ki67 expression was decreased in the inhibitor-treated group. (F) Relative fold expansion after inhibitor treatment. *P < .05; **P < .01. MFI, mean fluorescence intensity.
Inhibition of TRAF6 signaling impairs human tTreg expansion, Foxp3 expression and suppressive function. tTregs were treated with TRAF6 inhibitor for 2 days (n = 5). (A) Representative flow figures of FoxP3+ tTregs in different groups. Foxp3 expression was significantly decreased after inhibitor treatment. (B) FoxP3+ population and (C) FoxP3 expression was measured in these groups, and the inhibitor-treated group was significantly decreased. (D) CFSE assay was performed to measure suppressive ability and showed decreased suppressive function at 1:8 and 1:32. Values indicate mean ± SEM of these experiments. (E) Ki67 expression was measured in these groups, and Ki67 expression was decreased in the inhibitor-treated group. (F) Relative fold expansion after inhibitor treatment. *P < .05; **P < .01. MFI, mean fluorescence intensity.
Knockdown of miR-146b-5p increases nuclear localization of NF-κB, which is key in controlling tTreg function
TRAF6 is critical for TCR-mediated activation of NF-κB and acts by ubiquitinylating TAK1, leading to activation of IkBα kinase and destruction of inhibitor of NF-κBα (IκBα), which allows NF-κB to translocate to the nucleus.45,46 To determine whether miR-146 antagomir enhanced NF-κB nuclear translocation, in vitro–expanded tTregs were left untreated or treated for 2 days with scrambled RNA or miR-146b-5p antagomir; stained for CD4, NF-κB, and DRAQ5 (a nuclear marker); and analyzed using imaging flow cytometry.47 We acquired a similarity feature to quantify translocation; by this method, a more composited and similar image by 2 colors means more translocation to the nucleus. NF-κB (in yellow) and DRAQ5 (in red) are shown in Figure 5A; together with a composite image, we found more similar images in the antagomir group than the untreated/scramble group. Following data acquisition, the spatial relationship between NF-κB and DRAQ5 was measured using the similarity feature in the IDEAS software package (Figure 5B).48 Similarity scores measure the pixel intensity correlation between the anti–NF-κB and DRAQ5 images, and increased similarity scores indicate increased overlap. As shown in Figure 5C, antagomir-treated tTregs demonstrated significantly higher similarity scores than untreated or scramble-treated tTregs, indicating increased NF-κB nuclear localization. In conclusion, NF-κB localization is regulated by miR-146b-5p.
NF-κB activation is essential for human tTreg development and translocating NF-κB to the nucleus after knockdown of miR-146b-5p (n = 3). Cells were left untreated or were incubated with scramble RNA or miR-146b antagomir. Following treatment, cells were stained for CD4, NF-κB, and DRAQ5 and NF-κB nuclear localization determined by imaging flow cytometry. (A) Representative Imagestream images of cultured tTregs showing bright-field images, as well as individual or overlaid images of NF-κB and DRAQ5. (B-C) Representative (B) or summary (C) of similarity score measured by IDEA software quantitating the degree of overlap between NF-κB and DRAQ5 staining. Higher similarity scores indicate increased nuclear localization. For panels D-G, naive peripheral blood tTregs were purified, expanded in vitro, and were treated with either DMSO only or PS1145. (D) Representative example of Foxp3 vs CD127 staining on tTreg (gated on CD4+ cells). (E) Summary of overall %Foxp3+CD127− cells, and (F) level of Foxp3 expression. (G) CFSE assay for suppressive function in tTregs from each group. Values indicate mean ± SEM of these experiments. *P < .05; **P < .01. MFI, mean fluorescence intensity.
NF-κB activation is essential for human tTreg development and translocating NF-κB to the nucleus after knockdown of miR-146b-5p (n = 3). Cells were left untreated or were incubated with scramble RNA or miR-146b antagomir. Following treatment, cells were stained for CD4, NF-κB, and DRAQ5 and NF-κB nuclear localization determined by imaging flow cytometry. (A) Representative Imagestream images of cultured tTregs showing bright-field images, as well as individual or overlaid images of NF-κB and DRAQ5. (B-C) Representative (B) or summary (C) of similarity score measured by IDEA software quantitating the degree of overlap between NF-κB and DRAQ5 staining. Higher similarity scores indicate increased nuclear localization. For panels D-G, naive peripheral blood tTregs were purified, expanded in vitro, and were treated with either DMSO only or PS1145. (D) Representative example of Foxp3 vs CD127 staining on tTreg (gated on CD4+ cells). (E) Summary of overall %Foxp3+CD127− cells, and (F) level of Foxp3 expression. (G) CFSE assay for suppressive function in tTregs from each group. Values indicate mean ± SEM of these experiments. *P < .05; **P < .01. MFI, mean fluorescence intensity.
NF-κB activation is critical for the development of tTregs in mice and positively regulates FoxP3 expression.49,50 To address the role of NF-κB in human tTregs, an inhibitor of the NF-κB pathway (PS1145) was added for the final 2 days of culture.51 NF-κB inhibition decreased Foxp3 expression in a dose-dependent manner, leading to significantly decreased in vitro suppressive function (Figure 5D-G). Therefore, NF-κB nuclear localization is associated with human tTreg function.
Antagomir-treated tTregs show transcriptional signs of NF-κB activation
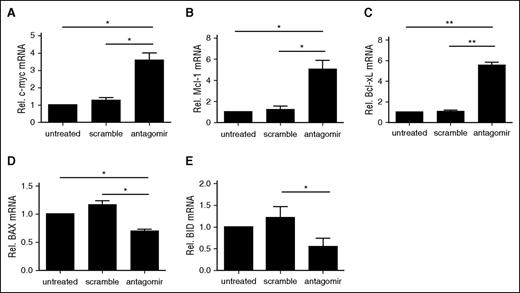
NF-κB activation in T cells results in a series of transcriptional changes that upregulates metabolism, cell cycle machinery, and prosurvival pathways. To determine whether the basal increase in NF-κB nuclear localization observed in antagomir-treated tTregs had transcriptional consequences, we compared the expression of known NF-κB–responsive genes in antagomir-treated tTregs with those left untreated or treated with scramble RNA. One NF-κB target gene, c-myc, is a crucial regulator of T-cell glycolysis and promotes T-cell activation–induced growth and proliferation.52,53 Thymocytes from miR-146b transgenic mice have defective IκB degradation following TCR stimulation and attenuated induction of c-myc.26,54 As shown in Figure 6A, antagomir treatment increased c-myc expression more than threefold over untreated or scramble-treated tTregs. In addition, consistent with the beneficial effects on viability and expansion, antagomir-treated tTregs had increased expression of antiapoptotic members of the Bcl-2 family (Bcl-xL and Mcl-1) and decreased expression of proapoptotic members (BID and BAX) (Figure 6B-F).
Treatment with miR-146b-5p antagomir increases antiapoptotic gene expression, decreases proapoptotic gene expression, and enhances tTreg persistence and expansion. Naive peripheral blood tTregs were sort-purified, expanded in vitro, and either left untreated or incubated with scramble RNA or miR-146b antagomir. Following treatment, RNA was purified and quantitative RT-PCR used to determine the expression of (A) c-Myc, the antiapoptotic genes (B) Bcl-xL and (C) Mcl-1, and the proapoptotic genes (D) BID and (E) BAX. Values indicate mean ± SEM of these experiments (*P < .05; **P < .01).
Treatment with miR-146b-5p antagomir increases antiapoptotic gene expression, decreases proapoptotic gene expression, and enhances tTreg persistence and expansion. Naive peripheral blood tTregs were sort-purified, expanded in vitro, and either left untreated or incubated with scramble RNA or miR-146b antagomir. Following treatment, RNA was purified and quantitative RT-PCR used to determine the expression of (A) c-Myc, the antiapoptotic genes (B) Bcl-xL and (C) Mcl-1, and the proapoptotic genes (D) BID and (E) BAX. Values indicate mean ± SEM of these experiments (*P < .05; **P < .01).
Together, these data show that miR-146b-5p antagomir treatment of tTregs results in NF-κB activation and expression of prosurvival genes that increase tTreg in vitro expansion and suppressive function.
miR-146-5p antagomir treatment significantly increases human tTreg efficacy and persistence in a xenogeneic model of GVHD
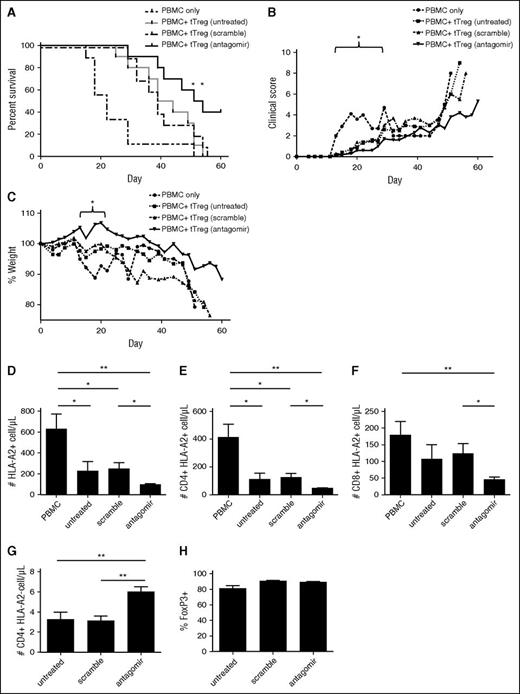
Because miR-146b knockdown enhances tTreg function, expansion, and survival in vitro, we sought to determine whether miR-146b-5p antagomir-treated tTregs would be more effective at preventing xenogeneic GVHD. Ex vivo–expanded tTregs (15 × 106) were untreated or treated with scramble or antagomir for 2 days, then injected with allogeneic PBMCs (15 × 106) into NSG mice. As shown in Figure 7A, all 3 groups of mice receiving tTregs had significantly reduced GVHD-induced lethality compared with PBMC-only controls (P < .05, .01, and .001 for untreated, scramble-, and antagomir-treated tTregs, respectively). Consistent with in vitro results, mice receiving antagomir-treated tTregs had significantly increased survival compared with mice receiving untreated or scramble-treated tTregs (P < .05 for both untreated and scramble-treated tTregs). Whereas no animals from the untreated tTreg control or scrambled groups survived past day 56, 40% receiving antagomir-treated tTregs survived beyond this day.
Antagomir-treated tTregs decrease mortality in a xenogeneic model of GVHD. Naive PB tTregs were sort-purified, expanded in vitro, and either left untreated or incubated with scramble RNA or miR-146b antagomir for 2 days. Following treatment, tTregs were washed and cotransferred (15 × 106) with allogeneic PBMCs (15 × 106) into NOD/Scid/γc−/− mice to assess the ability to ameliorate xenogeneic GVHD. n = 10, 10, 9 and 10 for the PBMC, untreated, scramble-treated, and antagomir-treated groups, respectively. (A) Kaplan-Meier survival curves for mice receiving PBMCs ± groups of tTregs. *P < .05. (B) Average weight (percentage of initial) for mice surviving on a given day for different groups of mice (*P < .05 for all tTreg groups from days 15 to 22). (C) Average GVHD score for mice surviving on a given day for different groups of mice. *P < .05 for all tTreg groups from days 13 to 22. GVHD severity was measured by enumerating PBMC-derived (ie, HLA-A2+) T-cell numbers in circulation on day 14 in the (D) HLA-A2+ total, (E) CD4+ HLA-A2+, and (F) CD8+ HLA-A2+ populations, respectively. (G) In vivo tTreg persistence was determined by enumerating CD4+ HLA-A2− cells in the blood on day 7. (H) The FoxP3+ tTreg population was maintained in all groups. Data shown are representative of 2 independent xenogeneic GVHD experiments.
Antagomir-treated tTregs decrease mortality in a xenogeneic model of GVHD. Naive PB tTregs were sort-purified, expanded in vitro, and either left untreated or incubated with scramble RNA or miR-146b antagomir for 2 days. Following treatment, tTregs were washed and cotransferred (15 × 106) with allogeneic PBMCs (15 × 106) into NOD/Scid/γc−/− mice to assess the ability to ameliorate xenogeneic GVHD. n = 10, 10, 9 and 10 for the PBMC, untreated, scramble-treated, and antagomir-treated groups, respectively. (A) Kaplan-Meier survival curves for mice receiving PBMCs ± groups of tTregs. *P < .05. (B) Average weight (percentage of initial) for mice surviving on a given day for different groups of mice (*P < .05 for all tTreg groups from days 15 to 22). (C) Average GVHD score for mice surviving on a given day for different groups of mice. *P < .05 for all tTreg groups from days 13 to 22. GVHD severity was measured by enumerating PBMC-derived (ie, HLA-A2+) T-cell numbers in circulation on day 14 in the (D) HLA-A2+ total, (E) CD4+ HLA-A2+, and (F) CD8+ HLA-A2+ populations, respectively. (G) In vivo tTreg persistence was determined by enumerating CD4+ HLA-A2− cells in the blood on day 7. (H) The FoxP3+ tTreg population was maintained in all groups. Data shown are representative of 2 independent xenogeneic GVHD experiments.
We have previously shown that adoptive transfer of tTregs ameliorates xenogeneic GVHD-associated pathologies, weight loss, clinical scores, and expansion of human T cells in peripheral blood (PB). All cohorts of tTreg-treated mice showed significantly decreased weight loss (P < .05) between days 15 and 22 and decreased clinical scores between days 13 and 22 (Figure 7B and Figure 7C, respectively). Although mice receiving antagomir-treated tTregs lost less weight and developed fewer clinical symptoms than mice receiving untreated or scramble-treated tTregs, these differences were not statistically significant.
Peripheral expansion of human T cells on days 14 to 20 correlates inversely with survival.55 To quantitate the ability of each Treg cohort to minimize expansion, HLA-A2 mismatching was used to distinguish GVHD-causing PBMCs (HLA-A2+) from tTregs (HLA-A2−). Mice were bled on day 14, and the total number of PBMC-derived cells per microliter of blood was enumerated (Figure 7D). To assess T-cell expansion, the number of CD4+HLA-A2+ and CD8+HLA-A2+ cells per microliter of blood was quantitated (Figure 7E-F). All mice receiving tTregs had reduced PBMC-derived CD4 T-cell numbers, and mice receiving antagomir-treated tTregs had significantly fewer CD4 T cells than mice receiving untreated or scramble-treated tTregs.
Previous studies found that tTreg persistence correlates with efficacy.55 We tested whether the enhanced survival of antagomir-treated tTregs seen in vitro might affect their persistence in vivo on days 7, 10, and 14. In both our xenogeneic GVHD model and in patients receiving third-party–expanded tTreg, we have shown that tTregs are difficult to detect in PB beyond days 10 to 12.7,55 Not surprisingly, the cell number on day 10 was hard to detect, and the number of tTregs observed on day 14 was not significantly higher than PBMC-only controls (which should have no HLA-A2+ cells) (data not shown). However, mice injected with miR-146b antagomir-treated tTregs had higher absolute numbers of circulating CD4+HLA-A2− cells on day 7 (Figure 7G) that mice in the scrambled or untreated groups (7.8 ± 3.2 cells/µL vs 4.6 ± 3.2 cells/µL vs 4.4 ± 3.5, P < .05 for miR-146b antagomir vs either group). Importantly, as observed in vitro (supplemental Figure 1B-D), antagomir-treated tTregs maintained a level of Foxp3 expression equivalent to untreated or scramble-treated tTregs (Figure 7H). Although increased PB tTreg number on day 7 is consistent with enhanced survival, other explanations include differences in expansion or homing.
Thus, knockdown of miR-146b-5p in tTregs increases in vivo efficacy and can be exploited to improve the efficacy of adoptive Treg therapy for the prevention of human GVHD.
Discussion
Our study suggests a new target to increase tTreg efficiency based on miRNA level. These observations include the following: (1) Both miR-146a and miR-146b-5p are highly differentially expressed in tTregs compared with control T-cells, but our results suggested that, in contrast to miR-146a, miR-146-5p negatively regulates FoxP3 expression, expansion. and tTreg function in vitro. (2) TRAF6, which plays an essential role in tTreg expansion and function, is a direct target of miRNA-146b-5p in human tTregs. (3) The NF-κB pathway is vital for FoxP3 maintenance, and miR-146b-5p antagomir–treated tTregs show enhanced nuclear localization of NF-κB. (4) Knockdown of miRNA-146b-5p prolongs tTreg survival by regulating NF-κB–related apoptosis/antiapoptosis genes. (5) Antagomir treatment enhances tTreg efficacy and persistence in a xenogeneic model of GVHD. While Bhairavabhotla et al did not find miR-146b antagomir affected Foxp3 expression or suppressive function, tTregs were stimulated with a suboptimal stimulus (plate-bound anti-CD3/28 vs artificial antigen-presenting cell) and antagomir treated at earlier time points in their studies (ie, days 1 and 3). In addition, Bhairavabhotla et al treated Tregs with PNA, which simultaneously reduced both miR-146a and miR-146b expression by >90%. Although some reports found miR-146a-5p positively regulates tTreg function in mice,25 we show that miR-146b-5p (alone) has a negative role controlling tTreg function. Therefore, it is possible that concurrent downmodulation of miR-146a and 146b has no net effect on Treg function. Based on these findings, we conclude that miR-146b-5p might be a therapeutic target for Treg clinical trials.
Although miR-146a and miR-146b-5p have similar overall sequences and share the same “seed regions,” they have different precursor miRNAs and distinct expression patterns and effects on target mRNAs.56-58 Previous studies have shown that inhibition of miR-146b-5p, but not miR-146a, significantly increased TRAF6 expression in human umbilical vein endothelial cells23 while inhibitors of miR-146a, but not miR-146b-5p, increased cyclooxygenase-2 protein abundance.59 Because miRNAs have many potential targets, we also cannot exclude the possibility that miR-146b binds to RNAs in Tregs that limit suppressive function, whereas in non-Tregs, miR-146b might have a different function by binding RNA partners that influence cellular functions other than suppression mechanisms. We have found that knockdown of miR-146b-5p favors FoxP3 expression and suppressive function in human tTregs, which indicates that miR-146b-5p and miR-146a play opposite roles. While miR-146a positively controls Foxp3 expression, overexpressed miR-146b-5p is linked to the pathogenesis of atherosclerosis with increased IL-17+CD4+ T-cell populations, suggesting that miR-146b-5p is more proinflammatory, leading to diminished Treg function.25,40,60 Combined with these studies, we think that the diametrically opposed functions of these miR-146 family members might be due to the need to maintain the proper Treg/Th17 balance in the human immune system to prevent autoimmune complications.
TRAF6 helps maintain FoxP3 expression and stability in Tregs. Although TRAF6 is a target of both miR-146a and miR-146b-5p in many cell types such as dendritic cells and cancer cells,19,43 knockdown of miR-146a increased tTreg TRAF6 expression in mice, but not human, tTregs31 and increased TRAF6 in murine tTregs were not responsible for enhanced tTreg function.25 We observed that miR-146b-5p directly targeted TRAF6 3′-UTR sites in human Tregs and that knockdown of miR-146b-5p enhanced FoxP3 expression and Treg function in vitro and in vivo. The importance of TRAF6 for tTreg expansion and function was confirmed using a TRAF6-specific inhibitor, which decreased Foxp3 expression, expansion, and suppressive function. In support of distinct roles for miR-146b-5p and miR-146a, knockdown of miR-146a did not alter TRAF6 expression in human tTregs. Thus, the miR-146b-5p–TRAF6 pathway, but not miR-146a pathways, control human tTreg function, expansion, and survival.
The NF-κB family of transcription factors is regulated by a family of inhibitors (IκB) that prevent nuclear translocation and by ubiquitin ligases that induce IκB degradation, resulting in NF-κB activation.61 Consistent with TRAF6 activation, miR-146b knockdown increased NF-κB nuclear localization in human tTregs, along with a concomitant increase in Foxp3 expression. NF-κB has also been shown to regulate several genes that are indispensable for tTreg survival, including antiapoptotic members (Bcl-2, Bcl-xL, and Mcl-1) and proapoptotic members (BID and BAX).62-65 Our experiments found that the enhanced in vitro and in vivo survival of miR-146b antagomir–treated tTregs correlated with increased expression of antiapoptosis genes (Bcl-xL and Mcl-1) and decreased expression of apoptosis genes (BID and BAX). Together, these results demonstrate that the miR-146b-5p–TRAF6–NF-κB pathway plays a critical role in regulating tTreg expansion, stability, and survival in vitro and in vivo (supplemental Figure 5).
miRNA-regulated pathways involve Treg function and survival. So far, 4 important pathways have been implicated: (1) miR-146a positively regulates Treg function through STAT125 ; (2) miR-99/150 could favor Treg differentiation by repressing the expression of the Th17-promoting factor mTOR39 ; (3) transferring miR-142-3p into Tregs impairs suppressive function by targeting AC9 mRNA33,35 ; and (4) although miR-155 doesn’t have an impact on FoxP3 expression in Treg, it does positively control Treg number in vivo, and inhibition of miR-155 sensitizes CD4+ Th cells to Treg-related suppression.66-69 Thus, up- or downregulation of specific miRNAs in Tregs could be a new therapy method used to treat GVHD patients in clinical trials. Here, we confirm another pathway miR-146b-5p–TRAF6–NF-κB could also contribute to this area.
tTreg adoptive cellular therapy has been shown to ameliorate autoimmune disease, graft rejection, and GVHD. Translating adoptive tTreg therapy to humans has proceeded slowly because of challenges in maintaining a high expression of Foxp3 in cultured cells, overall yield, and in vivo persistence of tTregs in GVHD patients.70,71 In this study, we have shown that nanoparticle-mediated delivery of miR-146b-5p antagomir to in vitro–expanded tTregs is an easy and efficient way of overcoming these challenges, which may be useful in clinical applications.
In summary, we demonstrate that a miR-146b-5p–TRAF6–NF-κB pathway plays an vital role in tTreg expansion, survival, and function in vivo and vitro. Focusing on miRNA might become a new starting point in Treg clinical trials.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported in part by research grants from the Children’s Cancer Research Fund (K.L.H. and B.R.B), Leukemia and Lymphoma Translational Research Grant R6029–07 (B.R.B.), National Institutes of Health (NIH) National Heart, Lung, and Blood Institute grant R01 HL11879 (B.R.B.), NIH National Cancer Institute (NCI) grant P01 CA067493 (B.R.B.), an NIH Clinical and Translational Science Award to the University of Minnesota (8UL1TR000114), and NIH/NCI grant P30 CA77598 utilizing the shared resource Flow Cytometry Core from the Masonic Cancer Center, University of Minnesota. This work was also supported in part by National Natural Science Fund grant 81571564 (L.L.), National Natural Science Fund: Outstanding Youth Fund grant 81522020 (L.L.), 863 Young Scientists Special Fund grant SS2015AA020932 (L.L.), and Major Program of the National Natural Science Foundation of China grant 91442117 (L.L.).
Authorship
Contribution: Y.L. designed and performed the experiments, evaluated the data, and wrote the manuscript; K.L.H. designed the experiments, evaluated the data, and wrote the manuscript; A.L.L., J.G., W.W., X.N., and P.R. performed the experiments and interpreted the data; B.L.L., J.L.R., and C.H.J. provided proprietary material and reviewed the manuscript; L.A.T. reviewed and discussed the data; D.H.M. discussed study design and interpretation of data and edited the manuscript; R.G. reviewed and discussed the data; and L.L. and B.R.B. designed the overall concept, analyzed the data, and wrote the manuscript.
Conflict-of-interest disclosure: B.R.B., B.L.L., J.L.R., and C.H.J. are founders of and scientific advisors for Tmunity Therapeutics and hold patents for the production and use of Tregs for clinical trials. The remaining authors declare no competing financial interests.
Correspondence: Ling Lu, Liver Transplantation Center, The First Affiliated Hospital of Nanjing Medical University, 300 Guangzhou Rd, 210029 Nanjing, China; e-mail: lvling@njmu.edu.cn; and Bruce R. Blazar, University of Minnesota, 420 Delaware St SE, Minneapolis, MN 55455; e-mail: blaza001@umn.edu.
References
Author notes
Y.L. and K.L.H. contributed equally to this study.
L.L. and B.R.B. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal