Key Points
SR-AI is the major receptor of FX at the macrophage surface.
Macrophages use SR-AI to control FX circulatory levels.
Abstract
Beside its classical role in the coagulation cascade, coagulation factor X (FX) is involved in several major biological processes including inflammation and enhancement of virus-induced immune responses. We recently reported that the long circulatory half-life of FX is linked to its interaction with liver-resident macrophages. Importantly, we now observed that macrophages, but not undifferentiated monocytes, support this interaction. Using cell biology approaches with primary and THP1-derived macrophages as well as transfected cells, we further identified the scavenger receptor type A member I (SR-AI) to be a macrophage-specific receptor for FX. This result was confirmed using SR-AI–deficient mice, which exhibit reduced circulating levels of FX in vivo and loss of FX-macrophage interactions in vitro. Binding studies using purified proteins revealed that FX binds specifically (half-maximal binding, 3 μg/mL) to the extracellular domain of SR-AI. Altogether, we demonstrate that macrophages regulate FX plasma levels in an SR-AI–dependent manner.
Introduction
Upon vessel damage, exposure of blood to collagen in the vessel wall and release of material by vascular cells triggers the activation of clotting factors. This provokes a series of events that eventually produces fibrin and leads to the formation of a hemostatic plug. During this process, called blood coagulation, zymogen factor X (FX) is converted into the activated serine protease FXa via site-specific proteolysis.1-5 The crucial role of FX in coagulation is demonstrated by the severe bleeding diathesis of patients with inherited FX deficiencies.6 However, beyond its role in coagulation, FX is also involved in nonhemostatic processes as first suggested by the partial embryonic lethality observed in mice following deletion of the F10 gene.7 Indeed, FX has been linked to fibroproliferative diseases, including tissue remodeling, fibrosis, cancer, and inflammation particularly after tissue injury in different pathologies.8,9 The mechanism by which FX modulates these processes is incompletely understood but seems to be related to the capacity of FXa to induce signaling pathways via the proteinase-activated receptors 1 and 2 (PAR-1 and -2). Another biological process involving FX concerns viral infections through its binding to the surface of herpes simplex virus-1 or human species C adenovirus. Whether such interactions promote the activation of the innate immune response10,11 or, in contrast, facilitate adenovirus and adenovirus-associated viruses infection12 is still a matter of debate. In any case, in view of this multiplicity of FX biological functions, further understanding of the regulatory mechanisms that control FX circulatory levels is clearly required.
Interestingly, the circulatory half-life of FX (48 hours) is longer than those of other structurally related coagulation factors such as FVII (5 hours), FIX (18-24 hours), or protein C (4 hours).13-15 However, the mechanisms underlying this particularly long half-life are still very poorly described. In a previous study conducted in our laboratory, organ biodistribution analysis identified the liver as a major target organ for FX.16 At the cellular level, we demonstrated that FX binds to Kupffer cells, the tissue-resident macrophages of the liver. Strikingly, we observed that macrophage inactivation in mice significantly reduced FX plasma levels. This was surprising because plasma concentrations of von Willebrand factor, a protein known to be degraded by macrophages, was increased under similar conditions.17 This strongly suggested that FX-macrophage interactions are crucial to the regulation of its circulating levels. However, the cellular receptor(s) and mechanism(s) involved in this interaction remained unknown.
In the present study, we aimed to explore the mechanism by which macrophages preserve FX plasma levels. To this end, we investigated the nature of the interaction between FX and macrophages and attempted to identify the cellular receptor(s) involved. Using multiple models both in vitro and in vivo, we identified the scavenger receptor type A member I (SR-AI) as a previously unreported receptor for FX and demonstrated its critical importance in maintaining normal FX plasma levels.
Methods
An extensive description of the experimental procedures can be found in the supplemental Methods (see supplemental Data, available on the Blood Web site), a brief summary of which is given below.
Proteins
Human plasma-derived FX was obtained from Cryopep, and was used throughout the study unless indicated otherwise. The soluble extracellular domains of human SR-AI (hSR-AI) and murine SR-AI (mSR-AI) were obtained from R&D Systems. hSR-AI was used throughout the study, unless stated otherwise.
Cell culture
Human macrophages were obtained both from THP1 (a monocytic leukemia cell line) and purified CD14+ cells. Murine macrophages were obtained from CD115+ cells.18 CD115+ cells were isolated by positive selection using the CD115+ sorting kit (Miltenyi Biotec). Differentiation into macrophages for each of these cells is described in more detail in the supplemental Methods. Complementary DNA encoding hSR-AI (Eurofins Scientific) was cloned and transfected into HEK-293 cells (see supplemental Methods).
Microscopy analyses and immunofluorescence-based quantification
Widefield-microscopy images were acquired on an AxioImager A1 (Carl Zeiss) and further analyzed using ImageJ software for quantification. Confocal microscopy images were acquired on a LSM700 (Carl Zeiss) and cell stacks were further deconvolved using Huygens software (Scientific Volume Imaging). The thresholded Manders coefficient (tMC) used to verify colocalization was calculated for each cell stack using the JACoP-plugin in ImageJ software. All images were assembled using ImageJ software (see supplemental Methods).
Flow cytometry
For flow cytometry analysis, THP1-derived macrophages or undifferentiated THP1 cells were incubated with Alexa 488–labeled FX or donkey anti-goat Fab′2 as a control as described in supplemental Methods. Event collection was performed using the Accuri C6 flow cytometer (BD Biosciences) and events were analyzed with Kaluza software (Beckman Coulter).
Binding assay
Binding of FX (0-40 μg/mL) to the soluble extracellular domain of hSR-AI (0.5 μg per well) in the presence or absence of potential competitors was assessed in an immunosorbent assay as described in the supplemental Methods.
FIX and FX activity assays
Endogenous murine FX activity was measured through FX activation into FXa by the Russel Viper Venom-X enzyme (RVV-X) and the subsequent conversion of the chromogenic substrate S-2765. FIX activity was determined using a 1-stage clotting assay.
Mice
Wild-type (wt) C57Bl/6 mice were purchased from Janvier Labs and SR-AI–deficient C57Bl/6 mice (B6.Cg-Msr1tm1Csk/J) were from The Jackson Laboratory. Blood samples were taken between 48 hours and 72 hours after arrival of the mice. Experiments using gadolinium chloride (GdCl3)-6H2O or polyclonal anti–mSR-AI antibodies were performed as described in the supplemental Methods.
Statistical analyses
Statistical analyses were performed using GraphPad Prism software. For flow cytometry data, a 2-way analysis of variance (ANOVA) followed by Sidak posttest for multiple comparisons was performed. Statistical significance of histograms was performed with Mann-Whitney nonparametric unpaired statistical tests. All graphs were built using GraphPad Prism software.
Results
FX binds to differentiated macrophages but not to monocytes
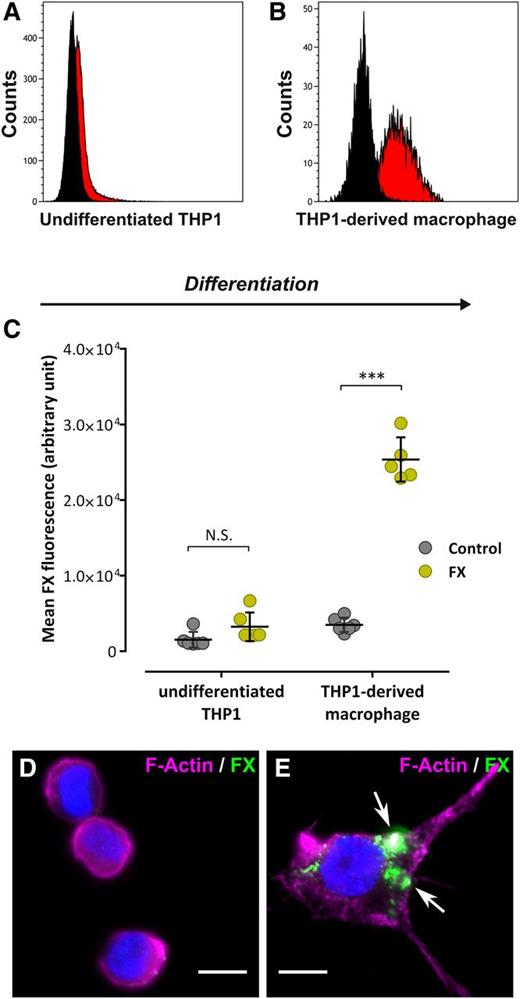
Recently, FX was found to interact with macrophages of different origin (Kupffer cells and THP1-derived macrophages).16 To explore the specificity of FX binding to macrophages vs monocytic cells, we compared binding of Alexa 488–labeled FX to undifferentiated THP1 cells and THP1-derived macrophages. Following incubation (1 hour at 37°C), flow cytometry analysis revealed a similar amount of fluorescence for undifferentiated THP1 cells incubated either with control Alexa 488–conjugated Fab′2 or Alexa 488–labeled FX (1.5 ± 1.0 × 103 and 3.2 ± 1.9 × 103 fluorescence units; P > .05), suggesting that FX is unable to bind these cells (Figure 1A,C). In contrast, THP1-derived macrophages incubated with Alexa 488–conjugated FX displayed a significant 10-fold shift of fluorescence compared with cells incubated with Alexa 488–conjugated Fab′2 (25.9 ± 2.9 × 103 and 3.0 ± 0.9 × 103 fluorescence units, respectively, P < .0001), indicating efficient binding of FX to these cells (Figure 1B-C). This result was confirmed using immunofluorescent staining (Figure 1D-E). No FX signal was visible using undifferentiated THP1 cells, whereas a cluster-shaped FX staining was observed for THP1-derived macrophages. Similar results were obtained using primary monocytes and monocyte-derived macrophages (data not shown). Binding of FX to THP1-derived macrophages but not undifferentiated THP1 cells indicates that candidate receptors for FX become available upon monocyte-to-macrophage differentiation.
FX binding to human monocytes and macrophages. (A-B) Undifferentiated THP1 (A) or THP1-derived macrophages (B) were incubated with Alexa 488–labeled FX or Alexa 488–labeled Fab′2 as a control (10 µg/mL, 1 hour at 37°C) and subsequently analyzed by flow cytometry for FX binding. Black curves represent Fab′2-incubated cells (control) whereas red curves represent Alexa 488-FX–incubated cells. Representative plots of 3 different experiments are shown. (C) The mean FX fluorescence was quantified and is expressed in arbitrary units. Each dot represents 1 experiment (N = 5-6 in total) and bars represent the mean ± SD. ***P < .001 in a 2-way ANOVA followed by the Sidak posttest for multiple comparison. (D-E) Widefield microscopy images of immunofluorescent staining for FX (green) in undifferentiated THP1 (D) or THP1-derived macrophages (E) incubated with 10 µg/mL FX (1 hour at 37°C). Nuclei and polymerized actin were counterstained using DAPI (blue) and Alexa 647–labeled phalloidin (magenta), respectively. Arrows indicate spots of FX staining. Bars represent 10 µm; objective, ×63. DAPI, 4,6 diamidino-2-phenylindole; N.S., not significant.
FX binding to human monocytes and macrophages. (A-B) Undifferentiated THP1 (A) or THP1-derived macrophages (B) were incubated with Alexa 488–labeled FX or Alexa 488–labeled Fab′2 as a control (10 µg/mL, 1 hour at 37°C) and subsequently analyzed by flow cytometry for FX binding. Black curves represent Fab′2-incubated cells (control) whereas red curves represent Alexa 488-FX–incubated cells. Representative plots of 3 different experiments are shown. (C) The mean FX fluorescence was quantified and is expressed in arbitrary units. Each dot represents 1 experiment (N = 5-6 in total) and bars represent the mean ± SD. ***P < .001 in a 2-way ANOVA followed by the Sidak posttest for multiple comparison. (D-E) Widefield microscopy images of immunofluorescent staining for FX (green) in undifferentiated THP1 (D) or THP1-derived macrophages (E) incubated with 10 µg/mL FX (1 hour at 37°C). Nuclei and polymerized actin were counterstained using DAPI (blue) and Alexa 647–labeled phalloidin (magenta), respectively. Arrows indicate spots of FX staining. Bars represent 10 µm; objective, ×63. DAPI, 4,6 diamidino-2-phenylindole; N.S., not significant.
FX colocalizes with SR-AI at the surface of THP1-derived macrophages
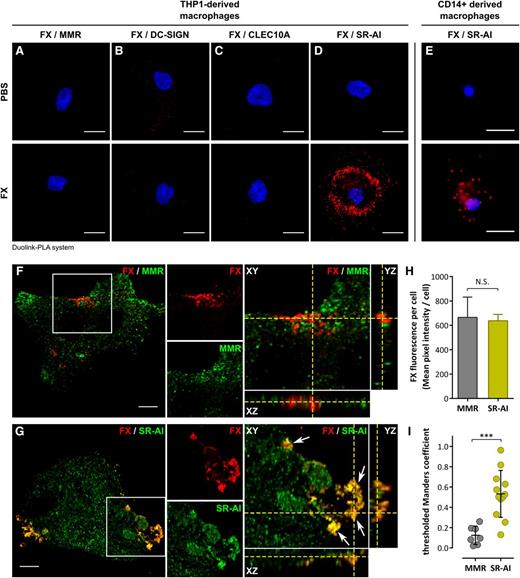
Analysis of literature and proteome databases revealed several transmembrane receptors upregulated upon monocyte-to-macrophage transition, 4 of which (mannose-macrophage receptor [MMR], dendritic cell-specific–intercellular adhesion molecule-3 grabbing nonintegrin [DC-SIGN], C-type lectin domain family 10 member A [CLEC10A], and SR-AI), were selected arbitrarily for testing their capacity to act as an FX receptor. We first performed Duolink–proximity ligation assay (PLA) analysis, which generates red fluorescent spots if proteins (in this case FX and 1 of the selected receptors) are located within a radius of 40 nm. No fluorescent spots were formed when cells were incubated with anti-FX and anti-receptor antibodies in the absence of FX (Figure 2A-E), confirming the specificity of this approach. A similar background signal was obtained for cells incubated with FX and stained for FX with MMR, DC-SIGN, or CLEC10A (Figure 2A-C), despite the presence of each of these receptors at the macrophage surface (supplemental Figure 1). In contrast, distinct red spots were detected for cells stained for FX and SR-AI (Figure 2D). This positive signal between FX and SR-AI was also detected using primary human CD14+ monocyte-derived macrophages (Figure 2E). Subsequent double immunofluorescent staining for FX and MMR or SR-AI was performed using THP1-derived macrophages incubated with FX and analyzed in confocal microscopy (Figure 2F-G). Little if any colocalization was detected for FX and MMR (Figure 2F). Signals for FX and SR-AI displayed similar staining patterns with clear areas of colocalization (yellow indicated with white arrows). Reconstituted orthoview (Figure 2G) shows colocalization between SR-AI and FX in all 3 planes (XY, XZ, and YZ) that seems to be restricted to the surface of macrophages. In order to verify this colocalization between FX and SR-AI, cell stacks acquired in confocal microscopy were analyzed to calculate the tMC. This coefficient (ranging from 0 to 1) is a statistical parameter allowing for the determination of whether 2 fluorescent signals overlap truly or coincidentally. To make this calculation, we compared cells stained for FX and MMR (negative control) or SR-AI. FX staining intensity was similar for both experiments (Figure 2H), ensuring that data analysis was unbiased by differences in FX binding between experiments. Macrophages stained for FX and MMR had a tMC value of 0.12 ± 0.09, whereas cells stained for FX and SR-AI had a tMC value of 0.53 ± 0.23 (mean ± standard deviation [SD]; P = .0005; Figure 2I). This statistical difference in tMC value confirms that the FX fluorescent signal truly overlaps with that of SR-AI.
Colocalization of FX in human macrophages. (A-E) Widefield microscopy images of Duolink-PLA assay between FX and MMR (A), DC-SIGN (B), CLEC10A (C), or SR-AI (D) in THP1-derived macrophages, or between FX and SR-AI in CD14+-derived macrophages (E) incubated with either PBS (top panels) or FX (bottom panels). Nuclei were counterstained using DAPI. Bars represent 10 µm; objective, ×40. Red spots indicate a distance <40 nm between 2 antigens. Images are representative of 3 different experiments. (F-I) Confocal analysis of immunofluorescent staining for FX (red) and MMR (F) or SR-AI (G) in a THP1-derived macrophage incubated with FX. Cell stack was reconstituted in orthoview to visualize colocalization of the 2 signals (right panels). Arrows indicate areas of colocalization. Z depth is 0.5 µm; bars represent 10 µm; objective, ×63. The mean FX fluorescence of the cells was quantified using Fiji software (H). tMC represents a statistical parameter verifying whether fluorescent signals truly overlap and was calculated using JACoP plugin in Fiji for THP1-derived macrophages incubated with FX and immunostained for FX and MMR (negative control) or SR-AI (I). ***P < .001, respectively, in the Mann-Whitney nonparametric unpaired statistical test. Dots represent each individual cell value and bars represent the mean ± SD of 7 (FX/MMR) to 12 (FX/SR-AI) cells from 3 independent experiments. PBS, phosphate-buffered saline.
Colocalization of FX in human macrophages. (A-E) Widefield microscopy images of Duolink-PLA assay between FX and MMR (A), DC-SIGN (B), CLEC10A (C), or SR-AI (D) in THP1-derived macrophages, or between FX and SR-AI in CD14+-derived macrophages (E) incubated with either PBS (top panels) or FX (bottom panels). Nuclei were counterstained using DAPI. Bars represent 10 µm; objective, ×40. Red spots indicate a distance <40 nm between 2 antigens. Images are representative of 3 different experiments. (F-I) Confocal analysis of immunofluorescent staining for FX (red) and MMR (F) or SR-AI (G) in a THP1-derived macrophage incubated with FX. Cell stack was reconstituted in orthoview to visualize colocalization of the 2 signals (right panels). Arrows indicate areas of colocalization. Z depth is 0.5 µm; bars represent 10 µm; objective, ×63. The mean FX fluorescence of the cells was quantified using Fiji software (H). tMC represents a statistical parameter verifying whether fluorescent signals truly overlap and was calculated using JACoP plugin in Fiji for THP1-derived macrophages incubated with FX and immunostained for FX and MMR (negative control) or SR-AI (I). ***P < .001, respectively, in the Mann-Whitney nonparametric unpaired statistical test. Dots represent each individual cell value and bars represent the mean ± SD of 7 (FX/MMR) to 12 (FX/SR-AI) cells from 3 independent experiments. PBS, phosphate-buffered saline.
FX binds in vitro to SR-AI
Having identified colocalization of FX with SR-AI at the macrophage surface, we then investigated binding of FX to the extracellular fragment of SR-AI in an immunosorbent assay using purified proteins. Little, if any, FX binding was detected to albumin-coated control wells. In contrast, FX binding to SR-AI was dose-dependent with a calculated half-maximal binding of 2.9 ± 0.9 µg/mL (Figure 3A). Subsequently, 3 potential competitors for SR-AI binding were tested. First, acetylated low-density lipoprotein (Ac-LDL; a natural ligand) failed to compete with FX (1 μg/mL) at concentrations up to 50 μg/mL, suggesting that both proteins bind to distinct regions within SR-AI. However, efficient dose-dependent inhibition was observed for polyinosinic acid (a polyanionic compound known to interact with SR-AI19 ) or polyclonal SR-AI–inhibiting antibodies (50% inhibitory concentration = 2.4 ± 0.1 µg/mL and 2.9 ± 0.2 µg/mL, respectively; Figure 3B). Polyinosinic acid also inhibited this interaction dose-dependently when tested at higher FX concentrations (5 μg/mL; supplemental Figure 2A). Binding was further reduced significantly in the presence of a soluble SR-AI fragment (supplemental Figure 2B). Thus, FX binds specifically to human SR-AI in a system using purified proteins.
Differential binding of FX and Ac-LDL to SR-AI. (A-B) Increasing concentration of FX (0-40 µg/mL) (A) or increasing concentration of SR-AI inhibitor (Poly[I], polyclonal anti–SR-AI antibody, or Ac-LDL; 1-50 µg/mL) along with 1 µg/mL FX (B) were incubated in microtiter wells coated with hSR-AI (0.5 μg per well). Bound FX was probed using a peroxidase-labeled polyclonal anti-FX antibody and revealed by chromogenic conversion of tetramethylbenzidine. For the negative control, hSR-AI was omitted during the coating (○ in panel A). Data represent the mean ± SD (n = 3-7). (C-H) Confocal analysis of THP1-derived macrophages incubated (1 hour at 37°C) with 150 nM Alexa 488–labeled Ac-LDL (C-D), 10 μg/mL Alexa 488–labeled FX (E-F), or preincubated with FX prior to the addition of Alexa 488–labeled Ac-LDL (G-H). Polymerized actin was counterstained using Alexa 647–labeled phalloidin (C,E,G) or cells were immunostained for EEA-1 (D,F,H). Dotted lines define cell boundaries based on phalloidin staining. Arrows indicate area of colocalization. Z depth is 0.5 µm; bars represent 10 µm; objective, ×63. OD, optical density; PoAb, polyclonal antibody; Poly[I], polyinosinic acid.
Differential binding of FX and Ac-LDL to SR-AI. (A-B) Increasing concentration of FX (0-40 µg/mL) (A) or increasing concentration of SR-AI inhibitor (Poly[I], polyclonal anti–SR-AI antibody, or Ac-LDL; 1-50 µg/mL) along with 1 µg/mL FX (B) were incubated in microtiter wells coated with hSR-AI (0.5 μg per well). Bound FX was probed using a peroxidase-labeled polyclonal anti-FX antibody and revealed by chromogenic conversion of tetramethylbenzidine. For the negative control, hSR-AI was omitted during the coating (○ in panel A). Data represent the mean ± SD (n = 3-7). (C-H) Confocal analysis of THP1-derived macrophages incubated (1 hour at 37°C) with 150 nM Alexa 488–labeled Ac-LDL (C-D), 10 μg/mL Alexa 488–labeled FX (E-F), or preincubated with FX prior to the addition of Alexa 488–labeled Ac-LDL (G-H). Polymerized actin was counterstained using Alexa 647–labeled phalloidin (C,E,G) or cells were immunostained for EEA-1 (D,F,H). Dotted lines define cell boundaries based on phalloidin staining. Arrows indicate area of colocalization. Z depth is 0.5 µm; bars represent 10 µm; objective, ×63. OD, optical density; PoAb, polyclonal antibody; Poly[I], polyinosinic acid.
Effect of FX on internalization of the SR-AI ligand Ac-LDL
We next compared binding and/or uptake of FX with the SR-AI ligand Ac-LDL by THP1 macrophages using confocal immunofluorescent microscopy. Alexa 488–conjugated Ac-LDL efficiently bound to THP1 macrophages, whereas costaining of the cytoskeleton using phalloidin revealed that Ac-LDL was located inside of the cell (Figure 3C). Indeed, additional analysis indicated that Ac-LDL colocalized with the early endosomal marker early endosome antigen-1 (EEA1) (Figure 3D). In contrast, costaining for FX and the cytoskeleton revealed that FX remained at the cell surface (Figure 3E). Indeed, no colocalization between FX and EEA1 could be detected (Figure 3F). Finally, we tested whether co-incubation of Ac-LDL with FX (at its plasma concentration of 10 μg/mL) would allow uptake of Ac-LDL. Confocal microscopical analysis showed that Ac-LDL was endocytosed by THP1 macrophages and delivered to the early endosomes in the presence of FX, whereas simultaneously FX remained at the cell surface (Figure 3G-H). Thus, despite the binding of FX to SR-AI, sufficient SR-AI receptors remain to mediate binding and uptake of other ligands for this receptor.
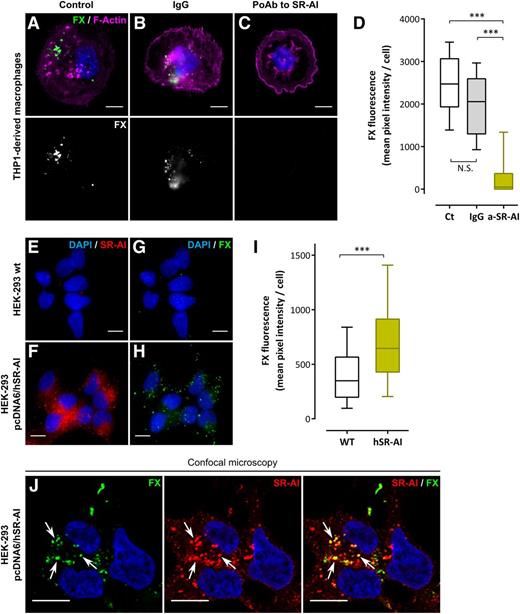
FX binds to SR-AI–transfected HEK-293 cells
To determine whether cellular SR-AI is able to act as a receptor for FX, binding of FX to THP1 macrophages was assessed in the presence of polyclonal anti–SR-AI antibodies. As depicted in Figure 4A-D, these antibodies reduced FX binding to THP1 macrophages by >80%. Additional binding experiments were performed using HEK-293 cells transfected or not with pcDNA6 encoding full-length hSR-AI (pcDNA6/hSR-AI). Transfection with pcDNA6/hSR-AI induced the abundant expression of SR-AI as assessed by immunofluorescent staining of nontransfected and transfected cells (Figure 4E-F). Following incubation with FX (1 hour at 37°C), nontransfected HEK-293 cells showed a weak background staining for FX (mean pixel intensity per cell: 419 ± 24 gray level units [GLU]; mean ± standard error of the mean [SEM]; n = 162 cells) (Figure 4G,I). In contrast, distinct bright spots of FX staining were observed with HEK-293 pcDNA6/hSR-AI cells incubated with FX (Figure 4H). Quantification revealed a significantly increased level of FX fluorescence (mean pixel intensity per cell: 728 ± 31 GLU; n = 218 cells; P < .0001) (Figure 4I). Moreover, HEK-293 pcDNA6/hSR-AI cells incubated with FX showed a similar staining pattern for SR-AI and FX with distinct areas of colocalization between the 2 signals when analyzed using confocal microscopy (white arrows in Figure 4J). Therefore, stable expression of SR-AI at the cell surface confers to the cell the ability to bind FX.
Binding of FX to cellular SR-AI. (A-D) THP1-derived macrophages were preincubated with PBS (A), nonspecific IgG (B), or a polyclonal anti–SR-AI antibody (C) and further incubated with 10 µg/mL Alexa 488–labeled FX (1 hour at 37°C). Images were acquired in widefield microscopy and quantified for FX fluorescence (D). (E-I) Immunofluorescent staining of SR-AI (red) and FX (green) was performed in nontransfected HEK-293 cells (E and G, respectively) or HEK-293 cells transfected with pcDNA6/hSR-AI (F and H, respectively) incubated with 10 µg/mL FX (1 hour at 37°C). Images were acquired in widefield microscopy and subsequently quantified for FX fluorescence (I). Data are presented in mean pixel intensity per cell (D,I). Boxes represent the median and 25th to 75th percentile, and bars represent the 10th to 90th percentile (at least 5 different fields per experiment in 3 different experiments). (J) Representative images of double immunostaining for FX and SR-AI in HEK-293 pcDNA6/hSR-AI analyzed using confocal microscopy. Nuclei and polymerized actin were counterstained using DAPI (blue) and Alexa 647–labeled phalloidin (magenta), respectively. Objective, ×63; bars represent 10 µm; Z depth is 0.4 µm (J) and arrows indicate area of colocalization. ***P < .001, respectively, in the Mann-Whitney nonparametric unpaired statistical test.
Binding of FX to cellular SR-AI. (A-D) THP1-derived macrophages were preincubated with PBS (A), nonspecific IgG (B), or a polyclonal anti–SR-AI antibody (C) and further incubated with 10 µg/mL Alexa 488–labeled FX (1 hour at 37°C). Images were acquired in widefield microscopy and quantified for FX fluorescence (D). (E-I) Immunofluorescent staining of SR-AI (red) and FX (green) was performed in nontransfected HEK-293 cells (E and G, respectively) or HEK-293 cells transfected with pcDNA6/hSR-AI (F and H, respectively) incubated with 10 µg/mL FX (1 hour at 37°C). Images were acquired in widefield microscopy and subsequently quantified for FX fluorescence (I). Data are presented in mean pixel intensity per cell (D,I). Boxes represent the median and 25th to 75th percentile, and bars represent the 10th to 90th percentile (at least 5 different fields per experiment in 3 different experiments). (J) Representative images of double immunostaining for FX and SR-AI in HEK-293 pcDNA6/hSR-AI analyzed using confocal microscopy. Nuclei and polymerized actin were counterstained using DAPI (blue) and Alexa 647–labeled phalloidin (magenta), respectively. Objective, ×63; bars represent 10 µm; Z depth is 0.4 µm (J) and arrows indicate area of colocalization. ***P < .001, respectively, in the Mann-Whitney nonparametric unpaired statistical test.
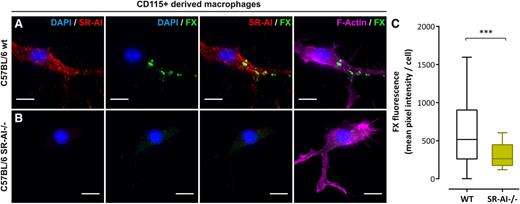
SR-AI–deficient macrophages lack FX binding
To deepen the physiological context of our findings, we included experiments using SR-AI–deficient C57BL/6 mice. First, CD115+ monocytes were purified from bone marrow of wt or SR-AI–deficient C57BL/6 mice and differentiated into macrophages. Immunostaining for mSR-AI confirmed its lack of expression in cells isolated from SR-AI–deficient mice (Figure 5A-B). After validating that human FX interacts with mSR-AI (supplemental Figure 3B), cells were incubated with human FX (10 µg/mL; 1 hour, 37°C). wt macrophages showed numerous areas of FX staining resembling those observed in human THP1-derived macrophages (Figure 5A). Quantification of FX fluorescence gave a mean pixel intensity of 654 ± 49 GLU (mean ± SEM; n = 97 cells; Figure 5C). However, FX staining was absent for mSR-AI–deficient macrophages (Figure 5B) and quantification evidenced a significant decrease in fluorescent signal (mean pixel intensity = 314 ± 20 GLU; n = 130 cells, P < .0001; Figure 5C). The absence of FX binding to SR-AI–deficient macrophages points to SR-AI being critical for the accumulation of FX at the macrophage surface.
FX binding to murine SR-AI–deficient macrophages. (A-C) CD115+-derived macrophages from wt (A) or SR-AI–deficient (B) C57BL/6 mice incubated with human FX (10 µg/mL, 1 hour at 37°C) were double immunostained for murine SR-AI (red) and human FX (green). Images were acquired in widefield microscopy and subsequently quantified for FX fluorescence (C). Data are presented in mean pixel intensity per cell. Boxes represent the median and 25th to 75th percentile, and bars represent the 10th to 90th percentile (at least 5 different fields per experiment in 3 different experiments). Nuclei and polymerized actin were counterstained using DAPI (blue) and Alexa 647–labeled phalloidin (magenta), respectively (A and B). Objective, ×63; bars represent 10 µm; ***P < .001, respectively, in the Mann-Whitney nonparametric unpaired statistical test.
FX binding to murine SR-AI–deficient macrophages. (A-C) CD115+-derived macrophages from wt (A) or SR-AI–deficient (B) C57BL/6 mice incubated with human FX (10 µg/mL, 1 hour at 37°C) were double immunostained for murine SR-AI (red) and human FX (green). Images were acquired in widefield microscopy and subsequently quantified for FX fluorescence (C). Data are presented in mean pixel intensity per cell. Boxes represent the median and 25th to 75th percentile, and bars represent the 10th to 90th percentile (at least 5 different fields per experiment in 3 different experiments). Nuclei and polymerized actin were counterstained using DAPI (blue) and Alexa 647–labeled phalloidin (magenta), respectively (A and B). Objective, ×63; bars represent 10 µm; ***P < .001, respectively, in the Mann-Whitney nonparametric unpaired statistical test.
Role of SR-AI in regulating FX plasma levels in vivo
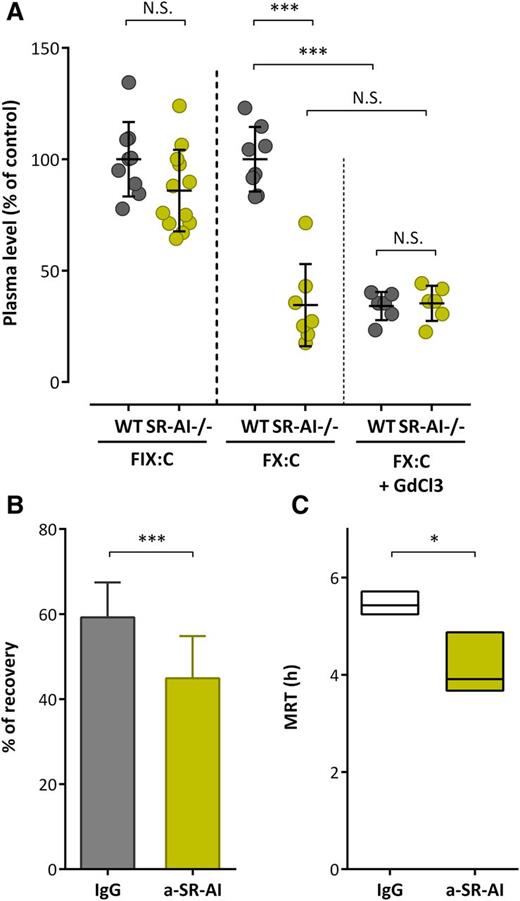
We then measured endogenous murine FIX and FX activity in order to investigate how the absence of mSR-AI affects FX plasma levels. When compared with wt C57BL/6 mice, SR-AI–deficient mice showed similar levels of mFIX (P > .05; Figure 6A). In contrast, a marked significant decrease in mFX activity was observed: 34.5% ± 18.4% of the wt value (mean ± SD; P < .0001; Figure 6A). To further explore the role of mSR-AI in vivo, we injected wt mice with polyclonal anti–mSR-AI or control antibodies (1.25 mg/kg) and mFX levels were measured 6 hours and 24 hours after antibody injection. Interestingly, endogenous mFX levels were reduced by 25% ± 9% and 40% ± 19%, respectively, upon anti–mSR-AI antibody injection compared with control (P < .01; supplemental Figure 3A). We then treated both wt- and SR-AI–deficient mice with GdCl3 (50 mg/kg), which inactivates macrophages. Endogenous mFX levels were decreased to 34% ± 6% 24 hours after GdCl3 application in wt mice (Figure 6A). In contrast, no further decrease in endogenous mFX levels was seen in SR-AI–deficient mice (35% ± 8% vs 34% ± 19% in GdCl3 and nontreated SR-AI–deficient mice, respectively; P > .05; Figure 6A). Finally, we tested the effect of inhibitory polyclonal anti–mSR-AI antibodies on the in vivo survival of IV administered human FX over a 24-hour period. Compared with control antibody-treated mice, the initial recovery of FX at 5 minutes after injection was significantly reduced in anti–mSR-AI–treated mice (59.2% ± 8.2% vs 44.9% ± 9.9%, for control and anti–SR-AI mice, respectively; n = 15-20 per group; P < .0001; Figure 6B). Furthermore, the mean residence time (MRT) was significantly shorter in anti–SR-AI–treated mice compared with control antibody-treated mice (MRT = 5.4 hours [minimum-maximum: 5.3-5.7 hours] vs 3.9 hours [minimum-maximum: 3.7-4.9 hours]; P < .05; Figure 6C). These data point to the absence of functional SR-AI being associated with a reduced circulatory half-life of FX, indicating that SR-AI is pertinent to sustain normal levels of FX.
SR-AI and FX levels in vivo. (A) Citrated plasma was collected from wt or SR-AI–deficient C57BL/6 mice prior or 24 hours after GdCl3 injection (50 mg/kg). FIX and FX activity were measured (A) and results are expressed in percentage of the normalized mean of wt values. (B-C) C57Bl/6 wt mice were injected in the caudal vein with FX (10 μg per mouse) in the presence of control IgG or polyclonal anti–mSR-AI antibodies (50 μg per mouse; 20 mouse per group). Plasma was collected at various time points (5 minutes, 1 hour, 3 hours, 6 hours, and 24 hours; 4-5 mice per group; 2-3 collections per mouse) and residual FX antigen was measured. Data were fitted to a biexponential decay equation to calculate pharmacokinetic parameters. Shown are the recovery at 5 minutes (B) and MRT (C) for each group. (A-B) Data represent the mean ± SD. (C) Boxes represent mean ± range of the calculated MRT, with the range being obtained from curves plotted with the minimum and maximum SD of residual FX levels. ***P < .001, respectively, in the Mann-Whitney nonparametric unpaired statistical test.
SR-AI and FX levels in vivo. (A) Citrated plasma was collected from wt or SR-AI–deficient C57BL/6 mice prior or 24 hours after GdCl3 injection (50 mg/kg). FIX and FX activity were measured (A) and results are expressed in percentage of the normalized mean of wt values. (B-C) C57Bl/6 wt mice were injected in the caudal vein with FX (10 μg per mouse) in the presence of control IgG or polyclonal anti–mSR-AI antibodies (50 μg per mouse; 20 mouse per group). Plasma was collected at various time points (5 minutes, 1 hour, 3 hours, 6 hours, and 24 hours; 4-5 mice per group; 2-3 collections per mouse) and residual FX antigen was measured. Data were fitted to a biexponential decay equation to calculate pharmacokinetic parameters. Shown are the recovery at 5 minutes (B) and MRT (C) for each group. (A-B) Data represent the mean ± SD. (C) Boxes represent mean ± range of the calculated MRT, with the range being obtained from curves plotted with the minimum and maximum SD of residual FX levels. ***P < .001, respectively, in the Mann-Whitney nonparametric unpaired statistical test.
Discussion
In this study, we have identified SR-AI as a receptor for FX on macrophages and revealed that their interaction is of physiological relevance to maintain normal FX plasma levels.
Previously, we have shown that FX is targeted to resident liver macrophages, and that GdCl3-mediated macrophage inactivation severely reduces FX plasma levels.16,20 Combined with the observation that FX remains at the surface of macrophages, rather than being internalized and degraded,16,20 these data point to macrophages serving a protective role in maintaining FX plasma levels. To better understand this protective role, we searched for receptors that are involved in the binding of FX to the macrophage surface. To limit the number of potential candidates, we first compared binding of FX to resting monocytes and differentiated macrophages. Interestingly, no binding of FX to resting monocytes was observed (Figure 1). Previously, Mac-1 has been proposed as a potential FX receptor at the surface of monocytes.21 However, these experiments were done using adenosine 5′-diphosphate– or ionomycin-stimulated monocytes, conditions that induce conformational changes within the Mac-1 integrin.22 The notion that resting monocytes were used in our experiments may explain the absence of FX binding to these cells (Figure 1). It should further be noted that treatment of mice with a Mac-1 inhibitor (ie, neutrophil-inhibitory factor) leaves FX levels unchanged (data not shown), suggesting that Mac-1 is not involved in the regulation of FX plasma levels.
A number of studies have investigated the changes in the transcriptome during monocyte-to-macrophage transition, revealing numerous receptors and other proteins that undergo important changes in expression during this transition.23-25 Based on these studies, we arbitrarily selected 4 candidates that could potentially act as receptor for FX. Three of them (MMR, DC-SIGN, and CLEC10A) belong to the C-type lectin receptors, characterized by their ability to bind carbohydrate structures in a calcium-dependent manner. MMR and DC-SIGN bind both mannose and fucose residues, whereas CLEC10A specifically recognizes α- and β-linked N-acetylgalactosamine.26-28 Each of these carbohydrate structures is present in FX,29 providing a rationale for a potential interaction between FX and these receptors. However, despite their evident expression in THP1 macrophages (supplemental Figure 1), no colocalization was found when assessed via Duolink-PLA analysis (Figure 2). Also, classic double staining did not reveal any colocalization between FX and any of these receptors, as is illustrated for MMR in Figure 2.
The fourth candidate was SR-AI, also known as macrophage-scavenger receptor or CD204. A first potential link between FX and SR-AI became apparent via Duolink-PLA analysis. This experiment revealed abundant colocalization of FX with SR-AI on both THP1- and primary CD14+-derived macrophages (Figure 2). Moreover, no binding of deglycosylated FX to macrophage-expressed SR-AI could be detected in this Duolink-PLA analysis (not shown), compatible with our previous finding that N-linked glycans on FX are needed for binding to macrophages.16 To confirm the close proximity between FX and SR-AI, classic double-staining experiments were performed using THP1 macrophages as well as stably transfected cells expressing SR-AI. In both cases, an apparent colocalization was observed when cells were analyzed via confocal microscopy (Figures 2G and 4J). Indeed, analysis of the fluorescent signals for FX and SR-AI on THP1 macrophages revealed a statistically relevant overlap of both signals (tMC = 0.53 compared with tMC = 0.12 for negative control; P < .0005; Figure 2I). Importantly, FX binding was reduced significantly to just above background levels, when analyzing binding of FX to murine SR-AI–deficient CD115+ macrophages (Figure 5). Moreover, binding of FX to THP1 macrophages was inhibited by >80% in the presence of anti–SR-AI antibodies (Figure 4A-D). These data point to SR-AI being the dominant receptor for FX on macrophages, although our findings do not exclude the presence of additional FX-binding receptors.
To further explore the interaction between FX and SR-AI, binding experiments using the soluble extracellular portion of SR-AI were performed. FX binding to immobilized SR-AI was saturable and dose-dependent, and half-maximal binding was calculated to be 3 μg/mL (Figure 3), thus allowing receptor binding at physiological plasma concentrations of FX (ie, 10 μg/mL). Polyclonal anti–SR-AI antibodies and polyinosinic acid were able to compete for FX binding in a dose-dependent manner in this system using purified proteins (Figure 3), indicating that FX binding to SR-AI is specific. Interestingly, binding was affected to a minor extent, if any, by the SR-AI ligand Ac-LDL. This lack of inhibition enabled us to test whether the presence of FX (which remains bound to SR-AI at the cell surface; Figure 3E-F) still allows the uptake of other SR-AI ligands, such as Ac-LDL. Confocal microscopy analysis clearly showed that FX did not prevent the binding and uptake of Ac-LDL by THP1 macrophages (Figure 3G-H), probably because of the large excess of SR-AI receptors over FX molecules at the macrophages surface.
It is intriguing to note that, to the best of our knowledge, FX is the first nonmodified circulating protein identified to interact with SR-AI. One could argue that purification of FX coincides with modifications in the protein that induce binding to SR-AI. However, we did not observe differences in SR-AI binding between plasma-derived FX or recombinant FX (not shown), both of which are purified via 2 different methods. Furthermore, FX levels are reduced by 65% in SR-AI–deficient mice. Should SR-AI binding require any FX modification, then one would not expect that >60% of the FX population in plasma would be dependent on regulation by SR-AI. Previously identified ligands included modified LDL variants, glycated albumin, glycated collagens, and β-amyloid fibrils.30 Such modified proteins are undesirable in the circulation, which may explain why they are efficiently eliminated from the circulation by SR-AI. For FX, a rather distinct mechanism appears to be involved. Indeed, far from being undesirable, appropriate plasma levels of FX are needed to sustain normal hemostasis. This may explain why other than “classic” ligands for SR-AI, FX is not endocytosed and degraded by macrophages even upon prolonged incubation (up to 2 hours).16 Unexpectedly, chemical depletion of macrophages using GdCl3 in wt mice results in FX levels that are decreased to about 35% of normal (Figure 6A), demonstrating that intact macrophages are needed to maintain FX plasma levels.16 Similarly, mice deficient for SR-AI are characterized by reduced FX levels (34% of normal; Figure 6A). These levels are no further decreased upon GdCl3 treatment in SR-AI–deficient mice (Figure 6A), suggesting that removal of SR-AI appeared responsible for the reduction of FX levels. Indeed, injection of anti–SR-AI antibodies significantly reduced FX levels in wt mice (supplemental Figure 3A). Moreover, the presence of SR-AI–blocking antibodies resulted in a significantly reduced recovery and MRT of human FX that was coadministered with these antibodies (Figure 6B-C). These data not only show that the interaction between FX and SR-AI is of high physiological significance, but also indicate that the FX/SR-AI interaction is fundamentally different compared with the interaction between SR-AI and other ligands. Apparently, SR-AI is critical in protecting FX in the circulation. This may seem counterintuitive in view of the functions and normal mode of action of macrophages in general and SR-AI in particular. However, this observation is not without precedent. Previously, carcinoembryonic antigen has been described as a glycoprotein that is recycled by macrophages rather than being degraded.31
Although our results show that SR-AI appears to play a prominent role in regulating plasma FX levels, so far we can only speculate on the reason why FX remains associated at the surface of macrophages in an SR-AI–dependent manner. It is tempting to consider a protein-recycling program resembling that of immunoglobulin G (IgG) and albumin, which via the neonatal Fc receptor are rescued from cellular catabolism.32,33 An essential difference is that the SR-AI/FX complex likely forms an extracellular reservoir, whereas IgG and albumin are recycled within the cells. Indeed, we have previously demonstrated that macrophage-bound FX is released in a time-dependent fashion from the macrophage surface as an intact, nonproteolyzed protein.16 The presence of such a reservoir would imply that circulating levels of FX in plasma represent only a part of the total FX population present in the vasculature. It should further be noted that our findings do not exclude the presence of other FX-binding receptors, and it seems likely that other partners at the macrophage surface contribute to the physiological role of SR-AI to maintain plasma levels of FX. Studies have been initiated to decipher this unusual mechanism that protects FX.
Apart from its importance to maintain FX plasma levels, the FX/SR-AI interaction at the macrophage surface may also be of relevance for FX functioning beyond hemostasis, for instance in relation to the innate immune system. Indeed, both FX and SR-AI contribute to the innate immune response via interactions with structural elements of viruses such as herpes simplex virus-1 and adenoviruses.10,11,34,35 In view of these observations, the possibility exists that the interaction between FX and SR-AI participates in the complex process of pathogen recognition and clearance, in which FX bridges SR-AI and the pathogen. Further studies are needed to decipher the precise role of the potential interaction between the FX/SR-AI complex and the innate immune system.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are grateful to Florence Fragnet and Anne-Lise Marville for their everyday skillful administrative assistance. The authors thank Pascal Roux and Dr Audrey Salles (Pasteur Institute, Paris, France) for their help in accessing the confocal microscope facility and interpretation of optical microscopy data, and Emilie Bouvier and Alexandre Diet (Center for Breeding & Distribution of Transgenic Animals, Orléans, France) for their technical assistance.
This work was supported by research funding from Novo Nordisk S/A. However, Novo Nordisk S/A had no role in the design of the study or the analyses and interpretation of the data.
Authorship
Contribution: V.M., A.B., C.L., A.H., and G.C. performed the experiments; V.M., C.V.D., P.J.L., and O.D.C. designed the study and analyzed the data; V.M., P.J.L., and O.D.C. wrote the paper; and all authors contributed to the editing of the final manuscript.
Conflict-of-interest disclosure: P.J.L. is a consultant for Novo Nordisk S/A. The remaining authors declare no competing financial interests.
Correspondence: Cécile V. Denis, INSERM U1176, 80 rue du General Leclerc, 94276 Le Kremlin-Bicêtre cedex, France; e-mail: cecile.denis@inserm.fr.
References
Author notes
P.J.L. and O.D.C. contributed equally to this manuscript.

![Figure 3. Differential binding of FX and Ac-LDL to SR-AI. (A-B) Increasing concentration of FX (0-40 µg/mL) (A) or increasing concentration of SR-AI inhibitor (Poly[I], polyclonal anti–SR-AI antibody, or Ac-LDL; 1-50 µg/mL) along with 1 µg/mL FX (B) were incubated in microtiter wells coated with hSR-AI (0.5 μg per well). Bound FX was probed using a peroxidase-labeled polyclonal anti-FX antibody and revealed by chromogenic conversion of tetramethylbenzidine. For the negative control, hSR-AI was omitted during the coating (○ in panel A). Data represent the mean ± SD (n = 3-7). (C-H) Confocal analysis of THP1-derived macrophages incubated (1 hour at 37°C) with 150 nM Alexa 488–labeled Ac-LDL (C-D), 10 μg/mL Alexa 488–labeled FX (E-F), or preincubated with FX prior to the addition of Alexa 488–labeled Ac-LDL (G-H). Polymerized actin was counterstained using Alexa 647–labeled phalloidin (C,E,G) or cells were immunostained for EEA-1 (D,F,H). Dotted lines define cell boundaries based on phalloidin staining. Arrows indicate area of colocalization. Z depth is 0.5 µm; bars represent 10 µm; objective, ×63. OD, optical density; PoAb, polyclonal antibody; Poly[I], polyinosinic acid.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/127/6/10.1182_blood-2015-05-647032/4/m_778f3.jpeg?Expires=1769088240&Signature=K~igwA5qPvkqeRHjPnWP2qNuDBoWW0OimjoU~H90OVnY551VRXFCp0661wX8ciAuyC9BeUx9OIeS6JOx-VkeV~7RZAnRWkY16D56fWka3EZ0edtLA~ck1hsE2F3OkfE669aDAG1qHFXk0M~xxjOkLFampr3fnjxZ0uwRKLaDjVLCxyZGTB3IVdh8Osrb0b5Z7yPEBpyAORPqL8kSNDIgFRfTi2JQKYV8Qr4LuvXwdMghKG5AucJHfEl1fTMeQzoc2Z7AEWu3zRccggq8m5kQsP0nOVfMw~87EZ-YI81JKFnaLPX4ksLA69-EKbWnNmic0JrHuEZZObxJoHMDYWiG4A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal