Key Points
A novel ELISA-based VWF multiplex activity assay assigns VWD phenotype among a cohort of type 1 and 2 VWD with an overall accuracy of >88%.
This assay shows correlation with traditional quantitative clinical VWF assays and may provide a rapid diagnostic method for variant VWD.
Abstract
Approximately 20% to 25% of patients with von Willebrand disease (VWD) have a qualitative defect of the von Willebrand factor (VWF) protein activities. Variant VWD typically is classified as type 1C, 2A, 2B, 2M, or 2N depending on the VWF activity defect. Traditionally, diagnosis has relied on multiple clinical laboratory assays to assign VWD phenotype. We developed an enzyme-linked immunosorbent assay (ELISA) to measure the various activities of VWF on a single plate and evaluated 160 patient samples enrolled in the Zimmerman Program for the Molecular and Clinical Biology of von Willebrand Disease with type 2 VWD. Using linear discriminate analysis (LDA), this assay was able to identify type 1C, 2A, 2B, 2M, or 2N VWD with an overall accuracy of 92.5% in the patient study cohort. LDA jackknife analysis, a statistical resampling technique, identified variant VWD with an overall accuracy of 88.1%, which predicts the assay’s performance in the general population. In addition, this assay demonstrated correlation with traditional clinical laboratory VWF assays. The VWF multiplex activity assay may be useful as a same-day screening assay when considering the diagnosis of variant VWD in an individual patient.
Medscape Continuing Medical Education online
This activity has been planned and implemented in accordance with the Essential Areas and policies of the Accreditation Council for Continuing Medical Education through the joint providership of Medscape, LLC and the American Society of Hematology.
Medscape, LLC is accredited by the ACCME to provide continuing medical education for physicians.
Medscape, LLC designates this Journal-based CME activity for a maximum of 1.0 AMA PRA Category 1 Credit(s)™. Physicians should claim only the credit commensurate with the extent of their participation in the activity.
All other clinicians completing this activity will be issued a certificate of participation. To participate in this journal CME activity: (1) review the learning objectives and author disclosures; (2) study the education content; (3) take the post-test with a 75% minimum passing score and complete the evaluation at http://www.medscape.org/journal/blood; and (4) view/print certificate. For CME questions, see page 2504.
Disclosures
Laurie Barclay, freelance writer and reviewer, Medscape, LLC, owns stock, stock options, or bonds from Pfizer. Associate Editor José López and the authors declare no competing financial interests.
Learning objectives
Evaluate the diagnostic performance of a new enzyme-linked immunosorbent assay (ELISA)-based von Willebrand factor (VWF) multiplex activity assay.
Determine the potential clinical usefulness of a new ELISA-based VWF multiplex activity assay.
Identify the individual components of a new ELISA-based VWF multiplex activity assay.
Release date: May 19, 2016; Expiration date: May 19, 2017
Introduction
Von Willebrand disease (VWD) is a common inherited bleeding disorder that may affect up to 0.1% of the population, manifesting as mucocutaneous bleeding, menorrhagia, and bleeding with surgical or other hemostatic challenges.1-6 Current VWD classification includes quantitative and qualitative defects.1 VWD type 1 is partial quantitative deficiency of von Willebrand factor (VWF), whereas VWD type 3 is complete deficiency of VWF.1 VWD type 1C, although not recognized in the current International Society on Thrombosis and Haemostasis classification, involves variant VWF with decreased survival or increased clearance and is determined, in part, by measuring relative concentration of VWF propeptide (VWFpp) compared with VWF antigen (VWF:Ag).7 VWD type 2A is decreased VWF-dependent platelet adhesion with deficiency of high-molecular-weight (HMW) VWF multimers because of abnormal synthesis or proteolysis.1 VWD type 2B involves increased affinity for platelet glycoprotein Ib (GPIb) resulting in increased VWF clearance, proteolysis, and usually thrombocytopenia.1 VWD type 2M is decreased VWF-dependent platelet adhesion via GPIb or collagen without significant loss of HMW multimers.1 VWD type 2N involves decreased VWF binding affinity for factor VIII (FVIII).1 Current phenotypic variant testing is cumbersome, performed by few specialized laboratories, and requires multiple VWF analyses. Thus, it may take weeks or longer to obtain definitive diagnosis.
New assays of VWF activities are essential to improve diagnostic fidelity and utility of VWD diagnosis. This is highlighted by the VWF ristocetin cofactor assay (VWF:RCo), which is a surrogate VWF activity test that has a high coefficient of variation (CV) up to 50% between laboratories8,9 and has impaired ristocetin-binding with a common VWF A1 domain single amino acid substitution, D1472H, in individuals without significant associated bleeding symptoms.10 To address these issues, development of ristocetin-free assays has been advocated.11,12 Flood et al were successful in developing a new assay of VWF GPIb activity using recombinant mutant GPIb on an enzyme-linked immunosorbent assay (ELISA) platform without requiring ristocetin (VWF:IbCo).10,13,14 Additionally, Patzke et al have described performance of the INNOVANCE VWF Ac (Siemens, Marburg, Germany) on the BCS XP System (Siemens) and the Sysmex CA-7000, CA-1500, CS-2000i, and CA-560 analyzers, which are also based on GPIb binding in the absence of ristocetin.15 Ristocetin-less assays, now collectively termed VWF:GPIbM,16 are steps toward improving the laboratory diagnosis of VWD. Additionally, clinically relevant VWF activity properties are being uncovered as described in collagen-VWF interactions.17-19 As additional, hemostatic VWF activities are revealed and impact on clinical VWD phenotype determined, unified VWF activity testing discriminating variant VWD is necessary.
Currently, assignment of type 2 VWD variants is highly variable among centers in the United States. Some centers define type 2 variants based on a semiarbitrary ratio of VWF:RCo/VWF:Ag 0.6 to 0.7.20-25 This is based on a ratio meant to identify patients with type 2A, 2B, and 2M VWD.24 However, type 2N VWD is not detectable with this approach and requires assessment of FVIII activity and VWF-FVIII binding.26,27 Moreover, not all VWD variants have the same clinical management; thus determining the precise variant of VWD is crucial to appropriate treatment. Typically, VWF activity tests are sent to specialty laboratories where assays are done for platelet binding, FVIII binding, collagen binding, or VWFpp, but tests may be sent to different laboratories. Furthermore, results may be based on different standards. Additionally, VWF genetic testing via exon 28 sequencing, where a majority of variant VWF sequence variations are found, or VWF full gene sequencing can be of clinical utility. Conceptually, a rapid, multicomponent assay providing a global approach to VWF biology discrimination and clinical VWD phenotype assignment using the same VWF standard would be desirable.
Based on this concept, we developed an ELISA-based VWF multiplex activity assay on plasma and applied this to subjects in the Zimmerman Program for the Molecular and Clinical Biology of von Willebrand Disease (ZPMCB-VWD). Data were analyzed with linear discriminant analysis (LDA), which gives optimal weights for tests to determine a variant VWD diagnostic algorithm. This VWF multiplex activity assay is qualitative and has potential to discriminate between types 2A, 2B, 2M, 2N, and 1C VWD in clinical laboratory settings.
Methods
Patient population
Patients were enrolled in ZPMCB-VWD through 8 primary centers in Milwaukee, Atlanta, Detroit, Huston, Indianapolis, Iowa City, New Orleans, and Pittsburgh, as well as secondary centers throughout the United States (see “Acknowledgments”). VWD subjects had preexisting diagnosis of VWD; however, final variant VWD diagnosis was assigned by ZPMCB-VWD based on current diagnostic guidelines1 and genotype data per the International Society on Thrombosis and Haemostasis Scientific and Standardization Committee VWF mutation database registry when available. VWF genotype of study subjects is available in the supplemental Data, available on the Blood Web site. Each center protocol was approved by its institutional Human Research Review Board, and all subjects gave informed consent in accordance with the Declaration of Helsinki.
One hundred thirty-four VWD, 22 hemophilia A (HA), and 4 type 3 VWD subjects were evaluated. HA subjects were enrolled from centers where VWD was suspected in families with known HA, carriers of HA, or subjects misdiagnosed as type 2N VWD. VWD subjects included in analysis were as follows: 21 type 1, 30 type 1C, 23 type 2A, 23 type 2B, 20 type 2M, and 17 type 2N. Samples were blinded to investigators.
VWF multiplex activity assay
Enrolled subjects’ blood was collected in 3.2% citrate, and plasma was prepared in a standard manner.10 Frozen plasma was sent to ZPMCB-VWD for evaluation. Monoclonal capture antibodies were used for VWF:Ag (AVW-5 and 105.4, 0.325 μg/mL), GPIbα (VWF:GPIbM/VWF:RCo, both 142.16, 5 μg/mL), FVIII (VWF:FVIIIB, 103.3, 1 mg/mL), and VWFpp (239.2 and 239.3, 0.325 μg/mL) coated in carbonate coating buffer (15 mmol/L sodium carbonate, 35 mmol/L sodium bicarbonate, 3 mmol/L sodium azide, pH 9.5), and collagen III (VWF:CB3, 3 μg/mL) was coated in 1× phosphate-buffered saline. Each individual VWF test was coated in its respective row on Immulon 4HBX (ThermoScientific) ELISA plates. Coated plates were premade by incubating at 4°C overnight. Three washes were used between each step. First, ELISA blocking buffer (1% bovine serum albumin in 1% phosphate-buffered saline, sterile-filtered) was incubated in all rows for 1 hour. Subsequently, a gain-of-function GPIb construct,13 50 U/dL in blocking buffer, was added for the VWF:GPIbM row; a wild-type GPIb construct,13 30 U/dL in blocking buffer, was added for the VWF:RCo row; recombinant FVIII (Kogenate, Berkeley, CA), 1 IU/mL in blocking buffer, was added for the VWF:FVIIIB row; blocking buffer was added to remaining rows; and the plate was incubated at room temperature for 1 hour. VWD study subject diluted plasma (1:50 in blocking buffer), diluted normal control plasma (1:50 in blocking buffer), or diluted 30% normal control plasma (normal control plasma diluted to 30% in type 3 VWD plasma, 1:50 in blocking buffer) was then added for each individual VWF test (effectively as a test “strip”) and incubated at room temperature for 1 hour. No ristocetin was used for VWF:GPIbM, but 1 mg/mL ristocetin was added for VWF:RCo assay. Subsequently, 2 biotinylated monoclonal anti-VWF antibodies, AVW-1 and AVW-15, were used to detect VWF (VWF:Ag, VWF:GPIbM, VWF:RCo, VWF:FVIIIB, and VWF:CB3), and 2 biotinylated monoclonal anti-VWFpp antibodies, 242.2 and 242.6, were used to detect VWFpp. Detection antibodies were 1 μg/mL antibody concentration, respectively, and were incubated at room temperature for 30 minutes. Streptavidin-conjugated alkaline phosphatase (1:5000 in blocking buffer) was then added and incubated for 30 minutes at room temperature. Finally, p-nitrophenyl phosphate (1:100 in substrate buffer, 0.1 g/L MgCl2 6H2O, 0.2 g/L NaN3, 1% diethanolamine, pH 9.8) was added and optical density (OD) measured on an ELISA plate reader at 405 to 650 nm after 30 minutes of room temperature incubation.
Analysis of VWF variants used the calculated ratio of the VWF activity test OD to VWF:Ag OD of the study subject. This ratio was then compared with the 30% normal control by taking the study subject’s OD ratio over the 30% normal control OD ratio for each VWF activity test. Standardization to 30% normal control plasma OD was chosen as opposed to 100% normal control plasma OD as the VWF:Ag in the majority of individuals with variant VWD is <50 IU/dL.1
ELISA variability studies
Variability studies were performed using lyophilized, reconstituted normal control and 30% normal control plasma. Normal control and 30% normal control plasma were aliquoted, frozen, and thawed on the day of testing. Twelve different runs were made on freshly prepared plates. Assays were performed on 4 different days to determine inter- and intraplate variability.
Statistical analysis
Statistics programs, SAS (SAS Institute Inc., Cary, NC) and SPSS (IBM, Armonk, NY), were used to perform analysis. To obtain a simultaneous prediction for each VWD phenotype, LDA was used as an algorithm to make phenotypic assignments. Addition of quadratic terms in the discriminant function accounted for nonlinearity in the data for a VWF phenotype.
A step-wise procedure for performing LDA is as follows: Complete linear discriminant function coefficients for each VWD phenotype and their quadratic terms were calculated based on the data. This gives a percentage predicted in each VWD category for each subject. The initial assumption is that there is no “a priori” preference for any of the VWD categories (this is the standard prior for LDA). Phenotype in the VWD categories is based on the LDA model score for each of the VWD categories.
The jackknife-resampling technique was used to validate LDA estimates and obtain confidence intervals for sensitivity and specificity of each type and overall for LDA. The jackknife-resampling technique predicts classification of a subject based on an LDA constructed without that observation and is far superior to validation by splitting the group into 2 parts (cross-validation).28 Furthermore, the jackknife method allows LDA data to be validated for application to a general population.
Additionally, individual VWF activity assay components were used as unique analytic discriminants between different VWD variants, and evaluated using the Mann-Whitney U test. Receiver operating characteristic (ROC) analysis was applied to determine optimal sensitivity and specificity to discriminate between VWD types.
VWF testing: for clinical comparison with VWF multiplex activity assay
Subject plasma was assayed using standard methods by the BloodCenter of Wisconsin Hemostasis Reference Laboratory for VWF:Ag, VWF:RCo, VWF:CB, VWFpp, VWF multimers, and FVIII activity as described previously.14 FVIII-binding activity was measured for type 2N VWD and HA subjects using plasma VWF bound to monoclonal antibody as previously reported.29 For subjects with suspected type 2B VWD, platelet-binding assays were performed using a modification of the “neutral” monoclonal antibody binding assay.30 ZPMCB-VWD variant VWD phenotypic assignment was made based on clinical laboratory data and VWF genotype analysis when available.
Results
VWF multiplex activity assay: ELISA consistency
Prior to evaluating the VWD study cohort, we examined the VWF multiplex activity assay for variability with normal control and 30% normal control plasma. Overall there was considerable consistency among measurements within a single ELISA plate. Intraplate variability was found to have a CV range of 4.7% to 6.2%, with median = 5.2%. Interplate variability was determined to have a CV range of 11.2% to 19.2%, with median = 14.1%. Thus, the assay provided consistent results within a plate, regardless of sample column allocation on a particular plate; however, assay consistency would need additional evaluation if implemented across various laboratories. Conceptually, this assay may be used in a “strip” format with samples run in single wells as opposed to duplicate or triplicate as done in standard quantitative clinical ELISA assays. In addition, plate-to-plate variability from coating the plate may be reduced through batch-produced precoated plates.
VWF multiplex activity assay: LDA
VWF activity profiles were generated for each subject and analyzed by LDA. LDA provided a unifying diagnostic algorithm and within this study cohort was able to correctly assign variant VWD phenotype in 124 of 134 (92.5%) subjects (Table 1). For example, 95.2% (n = 20) type 1 agreed between ZPMCB-VWD and LDA. Jackknife analysis applied to LDA correctly assigned variant VWD phenotype in 118 of 134 (88.1%) subjects (Table 1). Thus, this assay may correctly assign variant VWD phenotype 88.1% of the time in a population of subjects undergoing evaluation for “variant VWD.”
Overall LDA assignment of variant VWD phenotype
| . | LDA: number of observations and percent classified into VWD type . | ||||||
|---|---|---|---|---|---|---|---|
| Type 1 . | Type 1C . | Type 2A . | Type 2B . | Type 2M . | Type 2N . | Total by ZPMCB-VWD . | |
| ZPMCB-VWD confirmed VWD phenotype | |||||||
| VWD phenotype | |||||||
| Type 1 | 20 (19)* | 1 (2) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 21 |
| 95.24% (90.48%) | 4.76% (9.52%) | 0% (0%) | 0% (0%) | 0% (0%) | 0% (0%) | ||
| Type 1C | 2 (2) | 27 (26) | 1 (2) | 0 (0) | 0 (0) | 0 (0) | 30 |
| 6.67% (6.67%) | 90% (86.67%) | 3.33% (6.67%) | 0% (0%) | 0% (0%) | 0% (0%) | ||
| Type 2A | 0 (0) | 3 (3) | 19 (18) | 0 (0) | 1 (2) | 0 (0) | 23 |
| 0% (0%) | 13.04% (13.04%) | 82.61% (78.26%) | 0% (0%) | 4.35% (8.70%) | 0% (0%) | ||
| Type 2B | 0 (0) | 0 (0) | 0 (0) | 23 (23) | 0 (0) | 0 (0) | 23 |
| 0% (0%) | 0% (0%) | 0% (0%) | 100% (100%) | 0% (0%) | 0% (0%) | ||
| Type 2M | 1 (2) | 0 (0) | 0 (1) | 0 (0) | 19 (17) | 0 (0) | 20 |
| 5% (10%) | 0% (0%) | 0% (5%) | 0% (0%) | 95% (85%) | 0% (0%) | ||
| Type 2N | 1 (2) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 16 (15) | 17 |
| 5.88% (11.76%) | 0% (0%) | 0% (0%) | 0% (0%) | 0% (0%) | 94.12% (88.24%) | ||
| Total by LDA | 24 (25) | 31 (31) | 20 (21) | 23 (23) | 20 (19) | 16 (15) | 134 |
| . | LDA: number of observations and percent classified into VWD type . | ||||||
|---|---|---|---|---|---|---|---|
| Type 1 . | Type 1C . | Type 2A . | Type 2B . | Type 2M . | Type 2N . | Total by ZPMCB-VWD . | |
| ZPMCB-VWD confirmed VWD phenotype | |||||||
| VWD phenotype | |||||||
| Type 1 | 20 (19)* | 1 (2) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 21 |
| 95.24% (90.48%) | 4.76% (9.52%) | 0% (0%) | 0% (0%) | 0% (0%) | 0% (0%) | ||
| Type 1C | 2 (2) | 27 (26) | 1 (2) | 0 (0) | 0 (0) | 0 (0) | 30 |
| 6.67% (6.67%) | 90% (86.67%) | 3.33% (6.67%) | 0% (0%) | 0% (0%) | 0% (0%) | ||
| Type 2A | 0 (0) | 3 (3) | 19 (18) | 0 (0) | 1 (2) | 0 (0) | 23 |
| 0% (0%) | 13.04% (13.04%) | 82.61% (78.26%) | 0% (0%) | 4.35% (8.70%) | 0% (0%) | ||
| Type 2B | 0 (0) | 0 (0) | 0 (0) | 23 (23) | 0 (0) | 0 (0) | 23 |
| 0% (0%) | 0% (0%) | 0% (0%) | 100% (100%) | 0% (0%) | 0% (0%) | ||
| Type 2M | 1 (2) | 0 (0) | 0 (1) | 0 (0) | 19 (17) | 0 (0) | 20 |
| 5% (10%) | 0% (0%) | 0% (5%) | 0% (0%) | 95% (85%) | 0% (0%) | ||
| Type 2N | 1 (2) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 16 (15) | 17 |
| 5.88% (11.76%) | 0% (0%) | 0% (0%) | 0% (0%) | 0% (0%) | 94.12% (88.24%) | ||
| Total by LDA | 24 (25) | 31 (31) | 20 (21) | 23 (23) | 20 (19) | 16 (15) | 134 |
Columns depict LDA classification from diagnostic algorithm, and rows depict ZPMCB-VWD confirmed variant VWD diagnosis. The numerical value on the top of each row is the number of subjects LDA classified into each VWD variant phenotype, and the percent in the bottom of each row is the percent of ZPMCB-VWD confirmed diagnoses that LDA classified into each VWD variant. Rows equal 100%. Numerical values in parentheses depict jackknife-resampling analysis to predict assay performance in a general population.
Jackknife analysis.
To develop a clinically useful application, the probability of each variant VWD phenotype was determined for each study subject. This provides a clinically meaningful interpretation of most probable to least probable variant VWD diagnosis. Representative probability data are shown in Table 2. Complete probability data and coefficients for linear discriminant function can be found in the supplemental Data.
Variant VWD phenotype probability by LDA
| Subject ID . | Nonvariant . | Type 1C . | Type 2A . | Type 2B . | Type 2M . | Type 2N . |
|---|---|---|---|---|---|---|
| IN0270 | 0% | 5.4% | 94.6% | 0% | 0.01% | 0% |
| PB0169 | 7.3% | 92.5% | 0.08% | 0% | 0.13% | 0% |
| AT0003 | 0% | 0% | 0% | 100% | 0% | 0% |
| MK0260 | 0.57% | 2.1% | 1.8% | 0% | 95.5% | 0% |
| MK0039 | 1.2% | 2.5% | 0% | 0% | 0% | 96.3% |
| Subject ID . | Nonvariant . | Type 1C . | Type 2A . | Type 2B . | Type 2M . | Type 2N . |
|---|---|---|---|---|---|---|
| IN0270 | 0% | 5.4% | 94.6% | 0% | 0.01% | 0% |
| PB0169 | 7.3% | 92.5% | 0.08% | 0% | 0.13% | 0% |
| AT0003 | 0% | 0% | 0% | 100% | 0% | 0% |
| MK0260 | 0.57% | 2.1% | 1.8% | 0% | 95.5% | 0% |
| MK0039 | 1.2% | 2.5% | 0% | 0% | 0% | 96.3% |
Representative VWD phenotype probabilities (complete study cohort probability data available in the supplemental Data). Bolded percentage indicates ZPMCB-VWD confirmed diagnosis.
ROC curve discrimination between individual assay components
Table 3 depicts individual VWF activity discriminations, which are qualitatively descriptive of the biological differences seen in variant VWF activity. Of note, VWF:Ag ratio for type 1C having a lower ratio than type 2A may be significant only for this study population as both type 1C and 2A VWD subjects tend to have low VWF:Ag. Boxplots of these discriminations are available in the supplemental Data. ROC curves were used to evaluate individual VWF activity tests as individual discriminants and to demonstrate sensitivity vs specificity. ROC curves are shown in Figure 1. Interestingly, VWF:RCo was not found to have utility in discriminating among variant VWD phenotypes in this multiplex assay.
Comparison of VWF phenotypes between individual assay components
| Individual VWF activity test and variant VWD phenotype ratio comparison . | P . |
|---|---|
| VWF:GPIbM ratio [VWF:GPIbM/VWF:Ag]subject/[VWF:GPIbM/VWF:Ag]standard | |
| Type 2A ↓ ratio than type 1C | <.001 |
| Type 1 ↓ ratio than type 2B | <.001 |
| Type 2A ↓ ratio than type 2B | <.001 |
| VWF:FVIIIB ratio [VWF:FVIIIB/VWF:Ag]subject/[VWF:FVIIIB/VWF:Ag]standard | |
| Type 2N and 2N carriers ↓ ratio than HA (mild or carriers) | <.001 |
| VWF:CB3 ratio [VWF:CB3/VWF:Ag]subject/[VWF:CB3/VWF:Ag]standard | |
| Type 2A ↓ ratio than type 1C | <.001 |
| Type 2A ↓ ratio than type 2M | <.001 |
| VWFpp ratio [VWFpp/VWF:Ag]subject/[VWFpp/VWF:Ag]standard | |
| Type 2A ↓ ratio than type 1C | .002 |
| Type 1 ↓ ratio than type 1C | <.001 |
| VWF:Ag ratio [VWF:Ag]subject/[VWF:Ag]standard | |
| Type 1C ↓ ratio than type 2A | <.001 |
| Individual VWF activity test and variant VWD phenotype ratio comparison . | P . |
|---|---|
| VWF:GPIbM ratio [VWF:GPIbM/VWF:Ag]subject/[VWF:GPIbM/VWF:Ag]standard | |
| Type 2A ↓ ratio than type 1C | <.001 |
| Type 1 ↓ ratio than type 2B | <.001 |
| Type 2A ↓ ratio than type 2B | <.001 |
| VWF:FVIIIB ratio [VWF:FVIIIB/VWF:Ag]subject/[VWF:FVIIIB/VWF:Ag]standard | |
| Type 2N and 2N carriers ↓ ratio than HA (mild or carriers) | <.001 |
| VWF:CB3 ratio [VWF:CB3/VWF:Ag]subject/[VWF:CB3/VWF:Ag]standard | |
| Type 2A ↓ ratio than type 1C | <.001 |
| Type 2A ↓ ratio than type 2M | <.001 |
| VWFpp ratio [VWFpp/VWF:Ag]subject/[VWFpp/VWF:Ag]standard | |
| Type 2A ↓ ratio than type 1C | .002 |
| Type 1 ↓ ratio than type 1C | <.001 |
| VWF:Ag ratio [VWF:Ag]subject/[VWF:Ag]standard | |
| Type 1C ↓ ratio than type 2A | <.001 |
The ratio of each VWF activity test to VWF:Ag was evaluated for the ability to discriminate between VWF phenotypes.
ROC curve discrimination between individual assay components. Each ROC curve depicts individual VWF activity tests that together comprise the assay. ROC curve analysis shows each individual test’s ability to make VWF phenotype discriminations. (A) ROC curve: type 1C vs type 2A, VWF:GPIbM. (B) ROC curve: type 2B vs type 1, VWF:GPIbM. (C) ROC curve: type 2B vs type 2A, VWF:GPIbM. (D) ROC curve: HA (mild or carriers) vs type 2N, VWF:FVIIIB. (E) ROC curve: type 1C vs type 2A, VWF:CB3. (F) ROC curve: type 2M vs type 2A, VWF:CB3. (G) ROC curve: type 1C vs type 2A, VWFpp. (H) ROC curve: type 1C vs type 1, VWFpp. (I) ROC curve: type 2A vs type 1C, VWF:Ag. RT, ratio threshold.
ROC curve discrimination between individual assay components. Each ROC curve depicts individual VWF activity tests that together comprise the assay. ROC curve analysis shows each individual test’s ability to make VWF phenotype discriminations. (A) ROC curve: type 1C vs type 2A, VWF:GPIbM. (B) ROC curve: type 2B vs type 1, VWF:GPIbM. (C) ROC curve: type 2B vs type 2A, VWF:GPIbM. (D) ROC curve: HA (mild or carriers) vs type 2N, VWF:FVIIIB. (E) ROC curve: type 1C vs type 2A, VWF:CB3. (F) ROC curve: type 2M vs type 2A, VWF:CB3. (G) ROC curve: type 1C vs type 2A, VWFpp. (H) ROC curve: type 1C vs type 1, VWFpp. (I) ROC curve: type 2A vs type 1C, VWF:Ag. RT, ratio threshold.
VWF:GPIbM ratio as an individual discriminant.
VWF:GPIbM ratio discriminated type 1C from 2A with an ROC area under the curve (AUC) of 0.874 (95% CI, 0.780-0.968) (Figure 1A). VWF:GPIbM ratio at an RT >0.6366 (sensitivity 0.967, specificity 0.652) maximized separation of type 1C from 2A. VWF:GPIbM ratio discriminated type 2B from type 1 with an ROC AUC of 0.981 (95% CI, 0.951-1.000) (Figure 1B). VWF:GPIbM ratio at an RT >1.2676 (sensitivity 1.000, specificity 0.905) maximized separation of type 2B from type 1. VWF:GPIbM ratio discriminated type 2B from 2A with an ROC AUC of 1.000 (Figure 1C). VWF:GPIbM ratio at an RT of >1.1722 (sensitivity 1.000, specificity 1.000) maximized separation of type 2B from 2A.
VWF:FVIIIB ratio as an individual discriminant.
VWF:FVIIIB ratio discriminated HA (mild or carriers) from type 2N VWD with an ROC AUC of 1.000 (Figure 1D). VWF:FVIIIB ratio at an RT >0.8543 (sensitivity 1.000, specificity 1.000) maximized separation of HA (mild or carriers) from type 2N VWD.
VWF:CB3 ratio as an individual discriminant.
VWF:CB3 ratio discriminated type 1C from 2A with an ROC AUC of 0.922 (95% CI, 0.839-1.000) (Figure 1E). VWF:CB3 ratio at an RT >0.5971 (sensitivity 1.000, specificity 0.783) maximized separation of type 1C from 2A. VWF:CB3 ratio discriminated type 2M from 2A with an ROC AUC of 0.965 (95% CI, 0.916-1.000) (Figure 1F). VWF:CB3 ratio at an RT >0.6267 (sensitivity 1.000, specificity 0.826) maximized separation of type 2M from 2A.
VWFpp ratio as an individual discriminant.
VWFpp ratio discriminated type 1C from 2A with an ROC AUC of 0.757 (95% CI, 0.627-0.886) (Figure 1G). VWFpp ratio at an RT >2.5609 (sensitivity 0.633, specificity 0.783) maximized separation of type 1C from 2A. VWFpp ratio discriminated type 1C from type 1 with an ROC AUC of 0.965 (95% CI, 0.924-1.000) (Figure 1H). VWFpp ratio at an RT >2.2025 (sensitivity 0.800, specificity 1.000) maximized separation of type 1C from type 1.
VWF:Ag ratio as an individual discriminant.
VWF:Ag ratio discriminated type 2A from 1C with an ROC AUC of 0.867 (95% CI, 0.768-0.966) (Figure 1I). VWF:Ag ratio at an RT >0.9212 (sensitivity 0.696, specificity 0.933) maximized separation of type 2A from 1C.
VWF multiplex activity assay comparison with clinical data
Correlation with conventional assays of VWF physiological activity was made using data from ZPMCB-VWD obtained at the BloodCenter of Wisconsin clinical laboratory on the same plasma samples. Comparative analysis was performed on all available clinical data for the study cohort with clinical VWF:Ag, VWF:FVIIIB, VWFpp/VWF:Ag, VWF:CB3/VWF:Ag, and VWF:RCo/VWF:Ag.
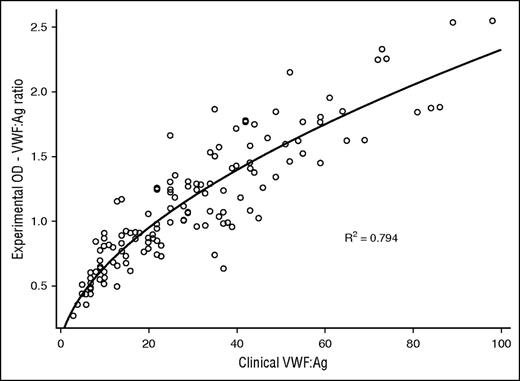
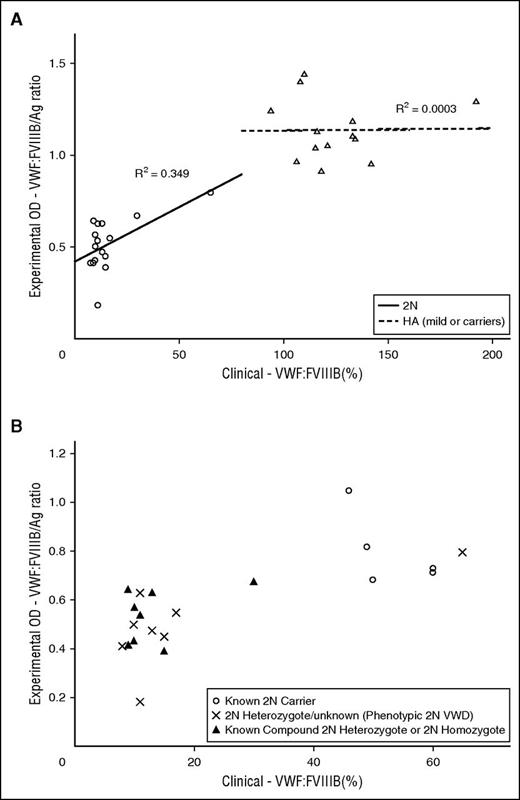
Evaluation of VWF:Ag of all VWD subjects demonstrated this assay correlated to the clinical assay, R2 = 0.794 (Figure 2). VWFpp analysis of 111 VWD subjects revealed correlation to the clinical assay, R2 = 0.630, and subanalysis of 51 type 1 and 1C VWD subjects showed R2 = 0.673. VWF:CB3 analysis of all type 2A, 2B, and 2M subjects revealed R2 = 0.675. VWF:CB3 analysis of 125 VWD subjects showed R2 = 0.527. VWF:GPIbM analysis was separated into type 2B and non-type-2B VWD subjects because of the significant VWF:GPIbM OD signal enhancement seen in this assay,13 which reflects augmented binding of type 2B VWF to GPIb. Type 2B subjects displayed VWF:GPIbM to clinical VWF:RCo correlation, R2 = 0.305, and 111 non-type-2B VWD subjects showed correlation with R2 = 0.429. VWF:FVIIIB comparison exhibited total separation of type 2N and HA, and VWF:FVIIIB R2 = 0.349 for type 2N subjects (Figure 3A), although there were only 16 type 2N subjects with clinical data available for analysis. There was also separation of type 2N VWD from type 2N carriers, although these data were not fit into a statistical model because there was paucity of subjects in each category (Figure 3B). Of note, there was 1 outlier phenotypic 2N VWD, heterozygote R854Q. This subject had a clinical VWF:RCo of 38 IU/dL and bleeding symptoms. Thus, the subject was classified as phenotypic type 2N, behaving clinically as a compound heterozygote. Additional figures of clinical correlation are available in supplemental Data.
Clinical VWF:FVIIIB vs experimental VWF:FVIIIB/Ag ratio. (A) Type 2N VWD and HA (mild or carriers) subjects. (B) Type 2N and 2N carrier VWD subjects. Type 2N VWD and type 2N carrier separation by clinical vs experimental VWF:FVIIIB assay. Type 2N carriers demonstrated both higher clinical and experimental VWF:FVIIIB than phenotypic 2N VWD or known compound 2N heterozygote or 2N homozygote subjects.
Clinical VWF:FVIIIB vs experimental VWF:FVIIIB/Ag ratio. (A) Type 2N VWD and HA (mild or carriers) subjects. (B) Type 2N and 2N carrier VWD subjects. Type 2N VWD and type 2N carrier separation by clinical vs experimental VWF:FVIIIB assay. Type 2N carriers demonstrated both higher clinical and experimental VWF:FVIIIB than phenotypic 2N VWD or known compound 2N heterozygote or 2N homozygote subjects.
Discussion
We describe an ELISA-based screening assay capable of providing a relatively comprehensive analysis of VWF physiological activity on a single testing platform using the same VWF standard. For an individual patient plasma sample, we are able to simultaneously analyze VWF binding activity to GPIb, collagen III, and FVIII, as well as measurement of VWF:Ag and VWFpp. When analyzed through LDA, our assay was able to correctly assign variant VWD patient phenotype into type 1C, 2A, 2B, 2M, or 2N with an overall accuracy of 92.5%. To predict how this assay might perform in the general population, jackknife analysis with LDA discriminated correct variant VWD phenotype assignment with an overall accuracy of 88.1%. Jackknife analysis gives the best estimate of accuracy for individuals within groups in general, rather than just this study cohort. This level of accuracy would be suitable for clinical application as a screening test. Providing a clinician with a percent probability of most to least likely variant VWD phenotype in an individual patient, as we have shown here (Table 3), would be desirable when needing to make timely patient management decisions, for example, identification of type 2B VWD when faced with the clinical decision to treat with desmopressin, which may cause increased thrombocytopenia in type 2B VWD.31 Another conceivable use would be to expand variant VWD testing on the commonly used ELISA platform to smaller, local hemostasis laboratories and potentially to reduce cost through multiple VWF activity tests done on a single platform.
New developments in laboratory evaluation for VWF over the past few years have been focused on replacing conventional VWF:RCo testing, because of its high CV and high limit of detection of 10 to 20 IU/dL,8,9,32,33 with new automated assays for VWF binding activity to GPIb; however, newer VWF:RCo methods have improved CV to 2% to 8% with automation.34 A few assays have taken advantage of GPIb binding independent of ristocetin through using recombinant human GPIb with 2 gain-of-function mutations inserted.12,13,35 Flood et al have described the VWF:IbCo assay, which demonstrated a CV of 7% and lower limit of detection of 3 U/dL.13 Lawrie et al have recently described a comparative analysis of the INNOVANCE VWF Ac relative to an established platelet-based VWF:RCo and report good correlation with VWF:Ac also having a lower limit of detection to 3 U/dL.35 These assays are performed on 2 different platforms with the VWF:Ac using polystyrene particles coated with anti-GPIb antibody35 and VWF:IbCo using an ELISA-based platform13 ; however, the former is not currently available for use in the United States. Additionally, other investigations have been done for VWF:GPIbM using similar in-house methods.36
Other modifications of the VWF:RCo assay have focused on improving automation on various photooptical coagulation analyzers to promote rapid turnaround times and more widespread availability. These assays have used chemiluminescense and turbidimetric detection.37-40 This current VWF multiplex activity assay takes advantage of incorporating the VWF:GPIbM assay13 and in this study was exclusively used to reflect VWF binding activity to GPIb in LDA and ROC analysis. Furthermore, in type 2B subjects, the VWF:GPIbM/VWF:Ag is significantly enhanced,13 which confers benefit of this by discriminating type 2B VWD from all other variant VWD. Incorporating the VWF:GPIbM also avoids misdiagnosing individuals with a common polymorphism, D1472H, as having type 2M VWD.10 VWF:RCo in our platform was not useful for discriminating variant VWD phenotypes, which is likely because of the well-known variability in that assay. Additionally, VWF:RCo in our assay requires the additional step of adding ristocetin, which is technically cumbersome. Therefore, moving forward, VWF:GPIbM could exclusively be used to measure GPIb binding activity in this assay.
ROC analysis with VWF:GPIbM ratio as an individual assay component was effective to discriminate type 1C from 2A, type 2B from type 1, and type 2B from 2A with an AUC >0.87 for all analyses. These properties reflect its overall contribution to variant VWD phenotype assignment in LDA. Although the correlation of VWF:GPIbM ratio to clinical VWF:RCo/VWF:Ag was lower than other VWF functional assay comparisons, this may be explained by inherent differences in the VWF:GPIbM and VWF:RCo assays.
VWF collagen binding (VWF:CB3) has long been discussed as a screening laboratory test to predict likelihood and probable subtype for diagnosing VWD41 and is also sensitive to the presence or absence of HMW multimers.42 Furthermore, VWF:CB can detect rare VWF mutations that exclusively affect collagen binding or variants with loss of HMW VWF multimers.43-45 VWF:CB ELISA assays performed in clinical laboratories typically use type I or III collagen, or a combination of both,46,47 and because of this we chose to include VWF:CB3 in this screening assay. ROC analysis with VWF:CB3 as an individual assay component showed it was effective to discriminate type 1C from 2A and type 2M from 2A with an AUC >0.92, which reflects its overall contribution to phenotype assignment in LDA. VWF:CB3 also showed correlation with clinical VWF:CB3/VWF:Ag with an R2 = 0.675. VWF binding to collagen I or III reflects VWF function in the A3 domain as compared with the A1 domain for VWF GPIb binding.48
VWF:FVIIIB being low is the necessary laboratory test to confirm type 2N VWD.26,49 This assay demonstrated efficient discrimination of HA from type 2N VWD with an ROC AUC = 1. This assay differs from commonly used VWF:FVIIIB platforms50 in that it detects VWF bound to FVIII rather than the converse and also has the ability to discriminate type 2N VWD, type 2N carriers, and HA as described in another VWF:FVIIIB ELISA.50
VWFpp is important for determining increased VWF clearance, and VWFpp or transient response to desmopressin is necessary for diagnosing type 1C VWD.7,51 This assay showed discrimination of type 1C from 2A, and also type 1C from type 1, with an ROC AUC = 0.965. In addition, VWFpp ratio showed correlation with clinical VWFpp/VWF:Ag with an R2 = 0.673 in type 1 and 1C subjects. Diagnosis of increased VWF clearance may be of critical importance because of implications for clinical management. For instance, a patient with increased VWF clearance may require VWF concentrate replacement rather than treatment with desmopressin in cases of severe bleeding.52
VWF:Ag measures the amount of protein present and has traditionally been performed by ELISA and latex immunoassay in standard practice.53,54 This assay provides a measurement of VWF:Ag compared with a 30% normal control and when run with a 100% normal control could provide a semiquantitative assessment of the test subject’s VWF:Ag level. It is also critical for all VWF functional ratios created by our assay, which allows for analysis by LDA. Additionally, our assay exhibited correlation with the clinical VWF:Ag with an R2 = 0.794, which is important when considering application as a clinical laboratory screening test.
Conceivably, this assay may be a more cost-effective approach for variant VWD laboratory evaluation on a single, widely used ELISA platform as compared with the multiple variant VWF tests currently employed, which cost multiple thousands of US dollars. However, this would require batch production of precoated ELISA plates rather than freshly coated plates as done in this study.
In summary, we describe a VWF multiplex activity assay that is able to discriminate variant VWD phenotypes 1C, 2A, 2B, 2M, and 2N on a single, ELISA platform that can provide same-day results. When analyzed through LDA, probability of each variant VWD phenotype can be determined. This assay qualitatively assesses VWF and demonstrates good correlation with traditional clinical laboratory quantitative tests. This assay method has potential for additional VWF physiological activities to be added, conceivably allowing for a more diverse investigation of VWF activities in an individual patient. Thus, this assay may be considered for screening patients for variant VWD with reasonable accuracy.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank all the subjects, physicians, and staff involved in the ZPMCB-VWD study, especially the directors of the primary clinical centers for the Zimmerman Program (see “Appendix”). Additional special thanks to Crystal Perry for her excellent assistance in sample and clinical data management.
In addition, numerous secondary centers contributed to subject recruitment: J. Hord, Akron Children’s Hospital, Akron, OH; M. Manco-Johnson, Mountain States Regional Hemophilia and Thrombosis Center, Aurora, CO; J. Strouse, Johns Hopkins Children’s Center, Baltimore, MD; A. Ma, University of North Carolina Chapel Hill, Chapel Hill, NC; L. Boggio and L. Valentino, Rush University Medical Center, Chicago, IL; R. Gruppo, Cincinnati Children’s Hospital, Cincinnati, OH; B. Kerlin, Nationwide Children’s Hospital, Columbus, OH; R. Kulkarni, Michigan State University, East Lansing, MI; D. Green, Northwestern University, Evanston, IL; D. Mahoney, Baylor College of Medicine, Houston, TX; A. Bedros, Loma Linda University Medical Center, Loma Linda, CA; C. Diamond, University of Wisconsin Madison, Madison, WI; A. Neff, Vanderbilt University, Nashville, TN; D. DiMichele and P. Giardina, Weill Cornell Medical College, New York, NY; A. Cohen, Newark Beth Israel Medical Center, Newark, NJ; E. Werner, Children’s Hospital of the King’s Daughters, Norfolk, VA; A. Matsunaga, Children’s Hospital & Research Center Oakland, Oakland, CA; M. Tarantino, Bleeding & Clotting Disorders Institute, Peoria, IL; F. Shafer, Drexel University College of Medicine, Philadelphia, PA; B. Konkle and A. Cuker, University of Pennsylvania, Philadelphia, PA; P. Kouides, Rochester General Hospital, Rochester, NY; D. Stein, Toledo Children’s Hospital, Toledo, OH; and A. Sharathkumar, Children’s Hospital of Chicago, Chicago, IL.
This work was supported by the grants from the National Institutes of Health National Heart, Lung, and Blood Institute (HL08158808 and HL112614) and the National Hemophilia Foundation NHF-Baxter Clinical Fellowship Grant.
Authorship
Contribution: J.C.R. and R.R.M. developed the assay; J.C.R., P.A.M., J.C.G., and R.R.M. designed the research; J.C.R. and P.A.C. collected data; J.C.R. performed experiments; K.Y. and R.G.H. performed the statistical analyses; J.C.R. and R.R.M. analyzed results and wrote the manuscript; and all authors edited the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
A complete list of the members of the Zimmerman Program Investigators appears in “Appendix.”
Correspondence: Jonathan C. Roberts, Bleeding & Clotting Disorders Institute, 9128 N. Lindbergh Dr, Peoria, IL 61615; e-mail: jroberts@ilbcdi.org.
Appendix: study group members
The Zimmerman Program Investigators are the directors of the primary clinical centers: T. Abshire, A. Dunn, and C. Bennett, Emory University School of Medicine, Atlanta, GA; J. Lusher and M. Rajpurkar, Wayne State University, Detroit, MI; D. Brown, University of Texas Health Science Center at Houston, Houston, TX; A. Shapiro, Indiana Hemophilia & Thrombosis Center, Indianapolis, IN; S. Lentz, University of Iowa, Iowa City, IA; J. Gill, Comprehensive Center for Bleeding Disorders, Milwaukee, WI; C. Leissinger, Tulane University Health Sciences Center, New Orleans, LA; and M. Ragni, University of Pittsburgh, Pittsburgh, PA.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal