Key Points
Cell-free (plasma) EBV DNA performs better than cellular EBV DNA as a marker of a broad range of EBV+ diseases.
Within a largely immunocompromised and hospitalized cohort, detection of EBV DNA in plasma is uncommon in the absence of EBV+ disease.
Abstract
Epstein-Barr virus (EBV) is a ubiquitous virus that establishes a latent infection within the host and in some cases can lead to the development of EBV-associated lymphomas, lymphoproliferative disorders, hemophagocytic lymphohistiocytosis, solid tumors, and other diseases. We studied the clinical significance of detecting EBV DNA in the plasma and peripheral blood mononuclear cells (PBMCs) of 2146 patients who had blood specimens sent to the Johns Hopkins Hospital clinical laboratory for viral quantitative real-time polymerase chain reaction assay over a 5-year period. Within this largely immunocompromised and hospitalized cohort, 535 patients (25%) had EBV detected in plasma or PBMCs. When EBV was detected in the absence of an EBV+ disease (n = 402), it was present only in PBMCs in 69% of cases. Immunocompromised patients were less likely to have EBV in plasma than in PBMCs in the absence of EBV+ disease. In patients with active, systemic EBV+ diseases (n = 105), EBV was detected in plasma in 99% of cases but detected in PBMCs in only 54%. Across a range of copy number cutoffs, EBV in plasma had higher specificity and sensitivity for EBV+ disease as compared with EBV in PBMCs. EBV copy number in plasma distinguished untreated, EBV+ lymphoma from EBV+ lymphoma in remission and EBV− lymphoma, and also distinguished untreated, EBV+ posttransplantation lymphoproliferative disorder (PTLD) from EBV+ PTLD in remission and EBV− PTLD. EBV copy number quantification is a useful diagnostic marker across the spectrum of EBV+ diseases, even among immunocompromised patients, with plasma specimens more indicative of EBV+ disease than PBMCs.
Introduction
Following primary Epstein-Barr virus (EBV) infection, which occurs in the vast majority of the world’s population, the virus establishes a latent reservoir in resting memory B lymphocytes. Although most people harbor EBV with no long-term clinical ramifications, EBV can be associated with a variety of hematologic diseases including Hodgkin lymphoma (HL), non-Hodgkin lymphoma (NHL), hemophagocytic lymphohistiocytosis (HLH), and posttransplantation lymphoproliferative disorder (PTLD), as well as solid tumors such as undifferentiated nasopharyngeal carcinoma (NPC). EBV DNA copy number quantification by real-time polymerase chain reaction (qPCR) of blood specimens is often used as a means of screening for these diseases or assessing treatment response. However, EBV DNA can be detected in the blood in the absence of an EBV-associated (EBV+) disease. Uncertainty remains regarding the significance of detecting EBV DNA in different blood compartments and of quantitative thresholds, particularly among acutely ill and/or immunocompromised patients.
Although EBV DNA qPCR assays are commonly used in the posttransplant setting,1-4 no consensus exists with regard to how to use EBV DNA in the blood to diagnose and/or manage EBV+ PTLD. Although elevated levels of EBV DNA in the blood of transplant recipients is associated with an increased risk for EBV+ PTLD and treatment responses are typically associated with decreases in EBV DNA copy number,5-7 the optimal blood compartment for these assessments and the copy number threshold that should trigger further clinical evaluation remains a matter of debate. Plasma, or plasma-containing specimens such as whole blood, appear more specific than peripheral blood mononuclear cell (PBMC) specimens for EBV+ PTLD, although the first harbinger of EBV+ PTLD is often rising levels of EBV DNA copies in PBMCs.8,9 However, EBV DNA copy number can frequently be elevated in the PBMCs of transplant patients during periods free of EBV+ PTLD, whereas EBV DNA in plasma seems to track better with disease activity.10,11
Outside the setting of transplant, the clinical significance of detecting EBV DNA in cellular vs cell-free blood compartments is even less well defined. Given the ubiquity of EBV infection, EBV DNA copy number in PBMCs or whole blood might serve as a general marker of immune function, which may have prognostic significance among patients with hematologic malignancies regardless of tumor EBV status.12 However, there are growing data to indicate that cell-free specimens are better than cellular specimens when using EBV DNA as a tumor marker. In a pilot study of 57 patients with EBV+ or EBV− malignancies, EBV DNA in whole blood was not a reliable marker of EBV+ tumors, suggesting that not all viral DNA in whole blood is tumor-related.13 By contrast, EBV DNA in plasma was a better marker than whole blood EBV DNA of EBV+ tumor response by radiologic assessment, indicating that it is likely that the cellular (PBMC) component impedes the use of whole blood EBV DNA as a tumor marker.13 Even patients infected with HIV+ have been shown to only rarely have EBV DNA detected in plasma or sera in the absence of EBV+ malignancy, whereas the detection of EBV DNA in PBMCs is more common.14,15
The assessment of EBV DNA in the blood continues to show promise as a clinically available, minimally invasive tumor marker across a range of malignancies. In NPC, cell-free EBV DNA is now well-established as a tumor marker, having proven useful in the diagnosis, staging, prognosis, and assessment of treatment response, often adding information about tumor status beyond that gleaned from radiologic, endoscopic, or surgical evaluations.16,17 As a tool for detecting subclinical EBV+ disease, one study demonstrated that the majority of deceased allogeneic bone marrow transplant patients with detectable cell-free EBV DNA were found to have PTLD at autopsy, whereas no patient with undetectable cell-free EBV DNA was diagnosed with PTLD post-mortem.18 As a prognostic marker in EBV+ hematologic malignancies, cell-free EBV DNA has been shown to be associated with survival outcomes in HL and extranodal natural killer/T-cell lymphoma (ENKTL), as well as complement or correlate with other blood-based prognostic markers, such as serum survivin in ENKTL and soluble CD163 in HL.19-23 Across a range of lymphomas, there is mounting evidence that the serial assessment of cell-free EBV DNA copy number can inform the dynamics of EBV+ disease status during and following therapy.24
Despite the growing interest in cell-free EBV DNA as a tumor marker, few studies have addressed the significance of detecting EBV DNA in hospitalized patients. In one study of plasma EBV DNA quantification among hospitalized patients and healthy controls, the frequency of EBV DNA detection corresponded to the severity of illness, where 32% of septic patients had EBV DNA detected in plasma, whereas only 5% of critically ill, nonseptic patients and 0.6% of healthy controls had EBV DNA detected in plasma.25 In a retrospective study of 62 hospitalized, nontransplant patients with detectable EBV DNA in plasma, the majority had lymphoid malignancies and/or was receiving T-cell–depleting agents, although the study only included 8 patients with EBV+ lymphoma and thus provided limited comparison of potential differences in copy number between those with and without EBV+ diseases.26 In HIV+ patients, EBV DNA was detected in the plasma more frequently and at higher levels in those with EBV+ lymphomas as compared with HIV+ patients with EBV− lymphomas and HIV+ patients without lymphoma but with opportunistic infections.27
The aim of this retrospective study was to determine the relative utility of EBV DNA quantification in plasma and PBMCs as a marker of EBV+ diseases among a large cohort of patients treated at Johns Hopkins Hospital for whom a specimen was sent for EBV DNA qPCR over a 5-year period.
Methods
Clinical record review
After Institutional Review Board approval, we retrospectively evaluated the clinical context in which patients treated at Johns Hopkins Hospital had specimens sent for EBV DNA quantification in plasma and PBMCs by qPCR between January 2007 and December 2011. For patients with an EBV+ disease and multiple specimens sent for qPCR, the specimen closest to the time of the diagnosis of the EBV+ disease and preceding any treatment or intervention was analyzed. For patients with no EBV+ diagnosis and multiple specimens, the specimen analyzed was either the first specimen where EBV DNA was detected or, if all specimens had no detectable EBV DNA, the first submitted specimen.
Patients were not eligible if there were no clinic notes that encompassed the specimen collection date. EBV+ disease was defined as a biopsy demonstrating EBV in tumor tissue or the presence of classic features and/or diagnostic criteria for an EBV+ disease (such as in infectious mononucleosis [IM] or HLH). Patients were considered to have unproven EBV+ PTLD if no diagnostic biopsy was performed but the patient received treatment of presumed PTLD (reduction of immunosuppression, rituximab, chemotherapy, virus-specific cytotoxic T lymphocytes, or other intervention). For patients with detectable EBV DNA, at least 1 year of clinical follow-up after specimen collection without a diagnosis of an EBV+ disease was required to meet the definition of no EBV+ disease. For all patients, information relating to immune status was collected, but detailed information regarding medical comorbidities was not collected given the large scope of the study.
Cases were grouped into 5 diagnostic categories: active, systemic (non-central nervous system [CNS]) EBV+ disease, EBV+ CNS disease, partially treated EBV+ disease, EBV+ disease in remission, or no EBV+ disease. The 9 active, systemic EBV+ disease groups were: IM, EBV+ HLH, biopsy-proven EBV+ PTLD, unproven EBV+ PTLD, EBV+ NHL, EBV+ HL, EBV+ lymphoproliferative disorder, EBV+ NPC, and oral hairy leukoplakia. The 2 EBV+ CNS diseases were: EBV encephalitis and EBV+ primary CNS lymphoma. Within the group of patients with no EBV+ disease, cases were further subdivided based on whether or not the patient was immunocompromised and the cause of immunocompromise (HIV, inherited, or pharmacologic).
Real-time qPCR
PBMCs were prepared by Ficoll-Hypaque density gradient separation. Automated counts (Sysmex KX-21N; Sysmex, Lincolnshire, IL) were performed to quantify recovered cells. DNA was isolated from PBMCs (2 × 106 cells) and plasma (200 μL) using QIAmp DNA blood mini reagents (Qiagen, Gaithersburg, MD). EBV qPCR was performed on DNA using Qiagen/Artus EBV analyte specific reagents (Qiagen) and 7500 Real-Time PCR System (Applied Biosystems/Thermo Fisher Scientific, Foster City, CA), with a 97 base pair region of EBV nuclear antigen-1 (EBNA-1) as the target. The limit of detection by qPCR was 50 copies per mL plasma or per 100 000 PBMCs.
Statistical analyses
Descriptive statistics were used to describe the cohort characteristics. Copy number distributions among groups were compared using the Kruskal–Wallis test with Dunn’s correction for multiple comparisons. Frequencies of EBV detection among groups were compared using the χ2 test. For correlations and regression analyses, EBV DNA copy numbers were log transformed. Correlations were computed using the Pearson test. Statistics were performed using GraphPad Prism version 6 (GraphPad software; La Jolla, CA) and Stata version 14 (StataCorp LP, College Station, TX). Two-sided P values of < .05 were considered statistically significant.
Results
Relationship between EBV DNA detection in plasma and/or PBMCs and EBV disease
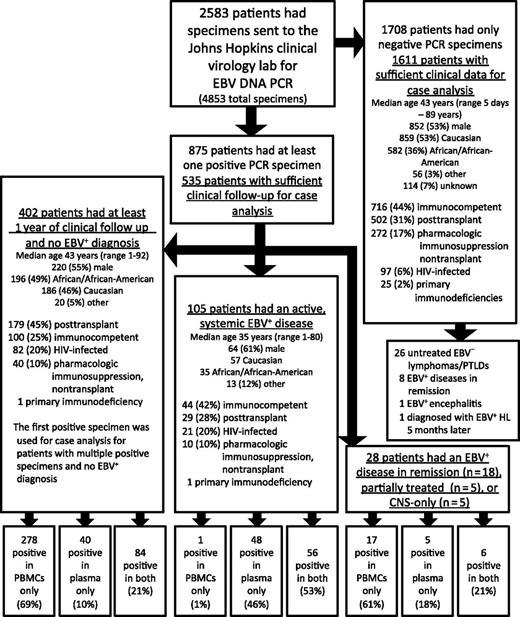
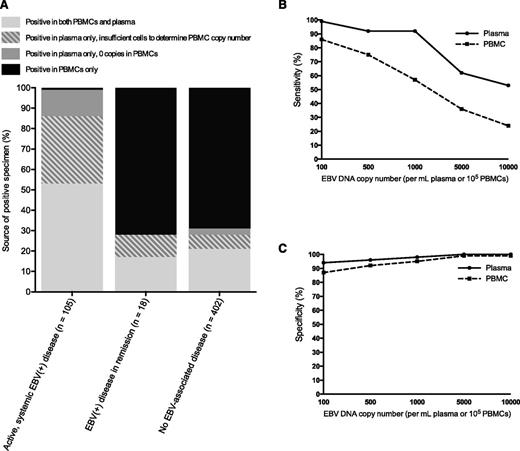
Of 2583 patients, 2146 patients (83%) had sufficient clinical information for case analysis, including 61% of patients with a positive specimen and 94% of patients with only negative specimens (Figure 1 and see annotated supplemental Figure 1, available on the Blood Web site). This difference in evaluable patients was due to the methodological requirement that those with a positive specimen had to have at least 1 year of clinical follow-up to try to exclude the presence of an evolving EBV+ disease in those with no apparent EBV+ disease at the time of qPCR. In the absence of an active, systemic EBV+ disease (n = 2041), EBV DNA was detected in 430 patients (21%), including 402 of 2004 patients (20%) with no prior, current, or subsequent EBV+ disease, 18 of 26 (69%) with EBV+ disease in remission, 5 of 5 (100%) with partially treated EBV+ disease, and 5 of 6 (83%) with CNS only EBV+ disease. Of the 2004 patients with no prior, current, or subsequent EBV+ disease, 18% had EBV DNA detected in PBMCs and 6% had EBV DNA detected in plasma (Table 1). The majority of patients with no EBV disease but a positive EBV specimen had EBV DNA detected in PBMCs only (n = 278 of 402, 69%) (Figure 2A). Among those with an active, systemic EBV+ disease (n = 105), 100% had EBV DNA detected in plasma or PBMCs and 104 (99%) had EBV DNA detected in plasma (Figure 2A). However, these patients less reliably had EBV DNA detected in PBMCs; of the 105, 13 (12%) had no EBV DNA detected in PBMCs, 35 (33%) had insufficient lymphocytes to determine copy number in PBMCs, and only 57 (54%) had EBV DNA detected in PBMCs at the time of diagnosis (McNemar χ2 10.29, P < .001). When positive specimens were associated with no EBV+ disease (n = 402) or EBV+ disease in remission (n = 18), the EBV DNA was detected in PBMCs only in the majority of patients (Figure 2A). Across a range of copy number cutoffs, EBV DNA in plasma had better sensitivity (Figure 2B) and specificity (Figure 2C) than PBMCs for distinguishing those with an active, systemic EBV+ disease from those with no prior, current, or subsequent history of an EBV+ disease. As many patients with EBV DNA in the blood were excluded due to insufficient follow-up, the above analyses were repeated to include the additional 219 patients with EBV in the blood and less follow-up, with the cutoff changed from 1 year to 1 week of clinical follow-up post-qPCR. The results of this re-analysis were similar, with PBMCs being the predominant source of a positive specimen in the absence of an EBV+ disease and plasma having a higher specificity than PBMCs for EBV+ disease across a range of cutoffs (supplemental Figure 2).
Flow diagram of patient specimens included in the study. Demographic information for each group is shown.
Flow diagram of patient specimens included in the study. Demographic information for each group is shown.
Detection of EBV DNA among patients with no current, prior, or subsequent EBV+ disease
| Diagnosis . | # of patients . | EBV DNA detected in plasma n (%) . | EBV DNA detected in PBMCs n (%) . | Median EBV DNA copies/mL plasma (range) . | Median EBV DNA copies/105 PBMCs (range) . |
|---|---|---|---|---|---|
| No EBV disease | 2004 | 121 (6) | 366 (18) | 0 (0-328 630) | 0 (0-76 414) |
| Immunocompetent | 808 | 28 (3) | 90 (11) | 0 (0-9966) | 0 (0-66 617) |
| Transplant recipient | 680 | 49 (7) | 161 (24) | 0 (0-328 630) | 0 (0-76 414) |
| On pharmacologic immunosuppression | 312 | 14 (4) | 37 (12) | 0 (0-3600) | 0 (0-2789) |
| HIV-infected | 178 | 29 (16) | 78 (44) | 0 (0-167 450) | 132 (0-34 433) |
| Primary immunodeficiency | 26 | 1 (4) | 0 (0) | 0 (0-70) | 0 (0-0) |
| Diagnosis . | # of patients . | EBV DNA detected in plasma n (%) . | EBV DNA detected in PBMCs n (%) . | Median EBV DNA copies/mL plasma (range) . | Median EBV DNA copies/105 PBMCs (range) . |
|---|---|---|---|---|---|
| No EBV disease | 2004 | 121 (6) | 366 (18) | 0 (0-328 630) | 0 (0-76 414) |
| Immunocompetent | 808 | 28 (3) | 90 (11) | 0 (0-9966) | 0 (0-66 617) |
| Transplant recipient | 680 | 49 (7) | 161 (24) | 0 (0-328 630) | 0 (0-76 414) |
| On pharmacologic immunosuppression | 312 | 14 (4) | 37 (12) | 0 (0-3600) | 0 (0-2789) |
| HIV-infected | 178 | 29 (16) | 78 (44) | 0 (0-167 450) | 132 (0-34 433) |
| Primary immunodeficiency | 26 | 1 (4) | 0 (0) | 0 (0-70) | 0 (0-0) |
Positive specimens by EBV disease status and test characteristics in plasma or PBMCs. (A) The percentage of patients with EBV DNA detected in plasma only, PBMCs only, or both plasma and PBMCs, separated by whether the patients had active, systemic EBV+ disease, EBV+ disease in remission, or no EBV+ disease. (B) For a range of cutoffs, the sensitivity of plasma (solid line) and PBMC (dashed line) specimens for distinguishing those with active, systemic EBV+ diseases from those with no EBV+ disease. (C) For a range of cutoffs, the specificity of plasma (solid line) and PBMC (dashed line) specimens for distinguishing those with active, systemic EBV+ diseases from those with no EBV+ disease.
Positive specimens by EBV disease status and test characteristics in plasma or PBMCs. (A) The percentage of patients with EBV DNA detected in plasma only, PBMCs only, or both plasma and PBMCs, separated by whether the patients had active, systemic EBV+ disease, EBV+ disease in remission, or no EBV+ disease. (B) For a range of cutoffs, the sensitivity of plasma (solid line) and PBMC (dashed line) specimens for distinguishing those with active, systemic EBV+ diseases from those with no EBV+ disease. (C) For a range of cutoffs, the specificity of plasma (solid line) and PBMC (dashed line) specimens for distinguishing those with active, systemic EBV+ diseases from those with no EBV+ disease.
Correlation between plasma and PBMC EBV DNA copy number
Of all specimens sent for EBV DNA qPCR (n = 4853), 1430 (29%) had insufficient cells to determine EBV DNA copy number in PBMCs. By contrast, EBV DNA quantification in plasma was possible for every specimen. Among specimens where EBV DNA could be quantified in both plasma and PBMCs (n = 3423), copy number was positively correlated among patients with active, systemic EBV+ disease (n = 70, Pearson r = 0.28), EBV+ disease in remission (n = 20, Pearson r = 0.65), and patients with no EBV disease (n = 1395, Pearson r = 0.45).
Negative specimens and relationship to disease
Of the 1611 patients with adequate clinical information for case analysis and no EBV DNA detected in the blood, none had active, systemic EBV+ disease diagnosed at the time of the negative specimen. Notably, however, 1 patient was subsequently diagnosed with EBV+ HL 5 months after the negative test, and had significant cervical and mediastinal lymphadenopathy concerning for lymphoma at the time of specimen collection. As the patient’s clinical history supports the likely presence of the EBV+ HL at the time of the negative specimen, the reason for this apparently false-negative EBV DNA qPCR result is not clear.
EBV DNA as a marker of active EBV+ disease
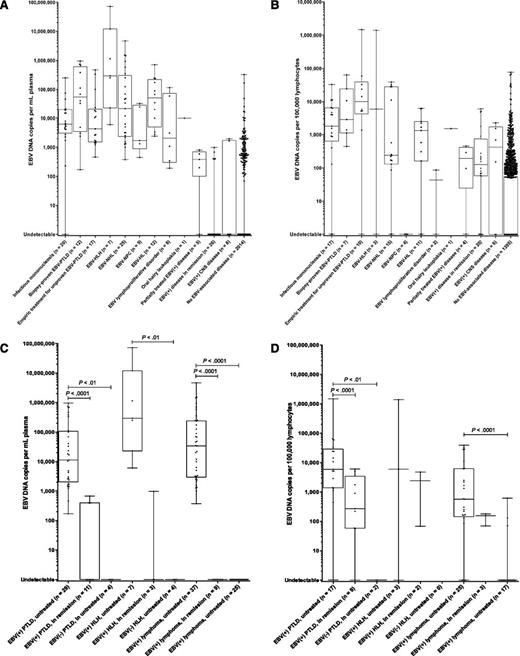
Over half of patients with an active, systemic EBV+ disease were immunocompromised, including 29 (28%) transplant recipients, 21 (20%) HIV+ patients, 10 (10%) patients receiving pharmacologic immunosuppression (nontransplant), and 1 patient with primary immunodeficiency. In plasma, EBV DNA copy number was higher among patients with active, systemic EBV+ disease (median, 14 783 copies/mL; interquartile range, 2554-106 449) compared with patients with EBV+ disease in remission (median, 0 copies/mL; interquartile range, 0-0) and patients with no EBV+ disease (median, 0 copies/mL; interquartile range, 0-0); Kruskal–Wallis P < .0001. Similar differences were observed in PBMCs, where patients with active, systemic EBV+ disease had a median of 1456 copies/105 PBMCs (interquartile range, 153-7673), as compared with those with EBV+ disease in remission (median, 126 copies/105 PBMCs; interquartile range, 58-743) and those with no EBV+ disease (median, 0 copies/105 PBMCs; interquartile range, 0-55); Kruskal–Wallis P < .0001. Descriptive statistics for EBV DNA copy number in plasma and PBMCs by disease group are shown in Table 2, with comparisons between the EBV+ disease groups (if the number of patients was ≥5) and patients with no EBV+ disease. In plasma and PBMCs, EBV DNA copy number was significantly higher in patients with IM, EBV+ PTLD, EBV+ NHL, and EBV+ HL as compared with those with no EBV+ disease. EBV DNA copy numbers in plasma and PBMCs by disease group are shown graphically in Figure 3A-B.
EBV DNA copy number in plasma and PBMCs by disease group
| Diagnosis . | # of patients with sufficient plasma for assay . | N (% of patients with EBV DNA in plasma) . | Median plasma copies/mL (range) . | Compared with patients with no EBV disease (n = 2014)* . | # of patients with sufficient PBMCs for assay . | N (% of patients with EBV DNA in PBMCs) . | Median PBMC copies / 100 000 lymphocytes (range) . | Compared with patients with no EBV disease (n = 1395)* . |
|---|---|---|---|---|---|---|---|---|
| Active and untreated, systemic EBV+ diseases | 105 | 104 (99) | 14 783 (0-7.3 × 107) | P < .0001 | 70 | 57 (81) | 1456 (0-1.5 × 106) | P < .0001 |
| IM | 20 | 19 (95) | 6273 (0-250 000) | P < .0001 | 17 | 15 (88) | 1793 (0-32 710) | P < .0001 |
| EBV+ HLH | 7 | 7 (100) | 295 720 (6054-7.3 × 107) | P < .001 | 3 | 2 (67) | 5998 (0-1.4 × 106) | — |
| Biopsy-proven EBV+ PTLD | 12 | 12 (100) | 54 960 (170-961 520) | P < .0001 | 7 | 7 (100) | 2842 (437-64 213) | P < .0001 |
| Empiric treatment of unproven EBV+ PTLD | 17 | 17 (100) | 4300 (460-471 100) | P < .0001 | 10 | 9 (90) | 10 085 (0-1.5 × 106) | P < .0001 |
| NPC | 5 | 5 (100) | 1690 (447-32 870) | P < .0001 | 4 | 0 (0) | 0 (0-0) | — |
| NHL | 24 | 24 (100) | 21 881 (372-4.7 × 106) | P < .0001 | 14 | 12 (86) | 239 (0-39 225) | P < .0001 |
| HIV-associated DLBCL | 7 | 7 (100) | 6300 (1290-1.4 × 106) | P < .001 | 3 | 3 (100) | 11 158 (170-39 225) | — |
| HIV plasmablastic lymphoma | 2 | 2 (100) | 7578 (3176-11 980) | — | 1 | 1 (100) | 28 072 | — |
| Peripheral T-cell lymphoma | 8 | 8 (100) | 47 445 (372-1.5 × 106) | P < .001 | 4 | 4 (100) | 14 355 (165-32 338) | — |
| ENKTL | 3 | 3 (100) | 935 764 (46 000-1.5 × 106) | — | 1 | 1 (100) | 32 338 | — |
| Peripheral T-cell lymphoma, other | 5 | 5 (100) | 5030 (372-125 988) | P < .01 | 3 | 3 (100) | 252 (165-28 458) | — |
| Immunodeficiency-associated DLBCL | 1 | 1 (100) | 70 277 | — | 1 | 0 (0) | 0 | — |
| Lymphomatoid granulomatosis | 4 | 4 (100) | 188 056 (2179-4.6 × 106) | — | 3 | 2 (67) | 221 (0-571) | — |
| Burkitt lymphoma | 2 | 2 (100) | 127 000 (1000-253 000) | — | 2 | 2 (100) | 108 (86-130) | — |
| HL | 12 | 12 (100) | 51 418 (2410-709 804) | P < .0001 | 11 | 9 (82) | 1328 (0-6319) | P < .0001 |
| HIV-associated | 9 | 9 (100) | 20 176 (2410-709 804) | P < .0001 | 8 | 7 (88) | 1498 (0-6319) | P < .0001 |
| Lymphoproliferative disorder | 7 | 7 (100) | 2076 (191-117 109) | P < .01 | 3 | 2 (67) | 86 (0-239) | — |
| Oral hairy leukoplakia | 1 | 1 (100) | 9901 | — | 1 | 1 (100) | 1517 | — |
| Partially-treated EBV+ disease | 5 | 4 (80) | 378 (0-805) | P ≤ .05 | 4 | 3 (75) | 192 (0-450) | — |
| EBV+ disease in remission | 26 | 5 (19) | 0 (0-980) | NS | 20 | 16 (80) | 126 (0-6053) | P < .0001 |
| EBV+ CNS disease | 6 | 2 (33) | 0 (0-1958) | NS | 6 | 4 (67) | 426 (0-2279) | P < .05 |
| Primary CNS lymphoma | 3 | 0 (0) | 0 (0-0) | — | 3 | 3 (100) | 700 (151-1549) | — |
| EBV encephalitis | 3 | 2 (67) | 1802 (1645-1958) | — | 3 | 1 (33) | 2279 | — |
| Diagnosis . | # of patients with sufficient plasma for assay . | N (% of patients with EBV DNA in plasma) . | Median plasma copies/mL (range) . | Compared with patients with no EBV disease (n = 2014)* . | # of patients with sufficient PBMCs for assay . | N (% of patients with EBV DNA in PBMCs) . | Median PBMC copies / 100 000 lymphocytes (range) . | Compared with patients with no EBV disease (n = 1395)* . |
|---|---|---|---|---|---|---|---|---|
| Active and untreated, systemic EBV+ diseases | 105 | 104 (99) | 14 783 (0-7.3 × 107) | P < .0001 | 70 | 57 (81) | 1456 (0-1.5 × 106) | P < .0001 |
| IM | 20 | 19 (95) | 6273 (0-250 000) | P < .0001 | 17 | 15 (88) | 1793 (0-32 710) | P < .0001 |
| EBV+ HLH | 7 | 7 (100) | 295 720 (6054-7.3 × 107) | P < .001 | 3 | 2 (67) | 5998 (0-1.4 × 106) | — |
| Biopsy-proven EBV+ PTLD | 12 | 12 (100) | 54 960 (170-961 520) | P < .0001 | 7 | 7 (100) | 2842 (437-64 213) | P < .0001 |
| Empiric treatment of unproven EBV+ PTLD | 17 | 17 (100) | 4300 (460-471 100) | P < .0001 | 10 | 9 (90) | 10 085 (0-1.5 × 106) | P < .0001 |
| NPC | 5 | 5 (100) | 1690 (447-32 870) | P < .0001 | 4 | 0 (0) | 0 (0-0) | — |
| NHL | 24 | 24 (100) | 21 881 (372-4.7 × 106) | P < .0001 | 14 | 12 (86) | 239 (0-39 225) | P < .0001 |
| HIV-associated DLBCL | 7 | 7 (100) | 6300 (1290-1.4 × 106) | P < .001 | 3 | 3 (100) | 11 158 (170-39 225) | — |
| HIV plasmablastic lymphoma | 2 | 2 (100) | 7578 (3176-11 980) | — | 1 | 1 (100) | 28 072 | — |
| Peripheral T-cell lymphoma | 8 | 8 (100) | 47 445 (372-1.5 × 106) | P < .001 | 4 | 4 (100) | 14 355 (165-32 338) | — |
| ENKTL | 3 | 3 (100) | 935 764 (46 000-1.5 × 106) | — | 1 | 1 (100) | 32 338 | — |
| Peripheral T-cell lymphoma, other | 5 | 5 (100) | 5030 (372-125 988) | P < .01 | 3 | 3 (100) | 252 (165-28 458) | — |
| Immunodeficiency-associated DLBCL | 1 | 1 (100) | 70 277 | — | 1 | 0 (0) | 0 | — |
| Lymphomatoid granulomatosis | 4 | 4 (100) | 188 056 (2179-4.6 × 106) | — | 3 | 2 (67) | 221 (0-571) | — |
| Burkitt lymphoma | 2 | 2 (100) | 127 000 (1000-253 000) | — | 2 | 2 (100) | 108 (86-130) | — |
| HL | 12 | 12 (100) | 51 418 (2410-709 804) | P < .0001 | 11 | 9 (82) | 1328 (0-6319) | P < .0001 |
| HIV-associated | 9 | 9 (100) | 20 176 (2410-709 804) | P < .0001 | 8 | 7 (88) | 1498 (0-6319) | P < .0001 |
| Lymphoproliferative disorder | 7 | 7 (100) | 2076 (191-117 109) | P < .01 | 3 | 2 (67) | 86 (0-239) | — |
| Oral hairy leukoplakia | 1 | 1 (100) | 9901 | — | 1 | 1 (100) | 1517 | — |
| Partially-treated EBV+ disease | 5 | 4 (80) | 378 (0-805) | P ≤ .05 | 4 | 3 (75) | 192 (0-450) | — |
| EBV+ disease in remission | 26 | 5 (19) | 0 (0-980) | NS | 20 | 16 (80) | 126 (0-6053) | P < .0001 |
| EBV+ CNS disease | 6 | 2 (33) | 0 (0-1958) | NS | 6 | 4 (67) | 426 (0-2279) | P < .05 |
| Primary CNS lymphoma | 3 | 0 (0) | 0 (0-0) | — | 3 | 3 (100) | 700 (151-1549) | — |
| EBV encephalitis | 3 | 2 (67) | 1802 (1645-1958) | — | 3 | 1 (33) | 2279 | — |
DLBCL, diffuse large B-cell lymphoma; NS, not significant.
Comparison by Kruskal–Wallis test, with Dunn’s correction for multiple comparisons.
Box-and-whisker plots of EBV DNA copy number by diagnosis and disease status. (A) EBV DNA copy number in plasma for patients with or without EBV+ diseases. (B) EBV DNA copy number in PBMCs for patients with or without EBV+ diseases. (C) Comparison of EBV DNA copy numbers in plasma of patients with PTLD, lymphoma, and HLH, by disease status and EBV status. (D) Comparison of EBV DNA copy numbers in PBMCs of patients with PTLD, lymphoma, and HLH, by disease status and EBV status. For all graphs, individual copy numbers (dots), median (line), 25th to 75th percentile (box), and range (whiskers) are shown for each patient group.
Box-and-whisker plots of EBV DNA copy number by diagnosis and disease status. (A) EBV DNA copy number in plasma for patients with or without EBV+ diseases. (B) EBV DNA copy number in PBMCs for patients with or without EBV+ diseases. (C) Comparison of EBV DNA copy numbers in plasma of patients with PTLD, lymphoma, and HLH, by disease status and EBV status. (D) Comparison of EBV DNA copy numbers in PBMCs of patients with PTLD, lymphoma, and HLH, by disease status and EBV status. For all graphs, individual copy numbers (dots), median (line), 25th to 75th percentile (box), and range (whiskers) are shown for each patient group.
Patients with active EBV+ lymphoma had higher EBV DNA copies in plasma than those with EBV+ lymphoma in remission or EBV− lymphoma, as did patients with active EBV+ PTLD when compared with those with EBV+ PTLD in remission or EBV− PTLD and patients with active EBV+ HLH as compared with EBV− HLH (Figure 3C). EBV DNA in PBMCs was a less reliable test for distinguishing these groups, in part because of insufficient cells to perform the assay for several patients, but could distinguish those with active EBV+ lymphoma from those with EBV− lymphomas, as well as those with active EBV+ PTLD from those with EBV+ PTLD in remission or EBV− PTLD (Figure 3D).
Detection of EBV DNA in blood among patients with no EBV+ disease
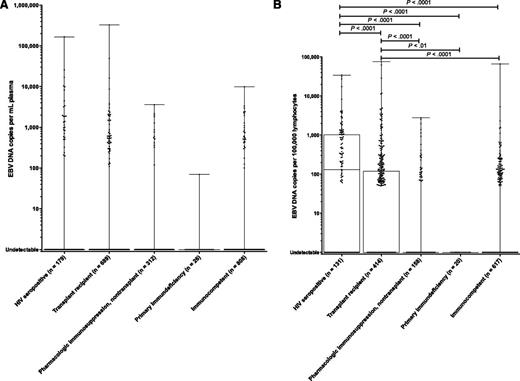
Of the patients (n = 402) who had EBV DNA detected in PBMCs, plasma, or both but no EBV+ diagnosis, the median follow-up without a subsequent EBV+ diagnosis was 3.5 years beyond the positive specimen (range, 1.0-7.6 years). The majority of these patients were immunocompromised (n = 302, 75%); 179 (45%) were transplant recipients, 82 (20%) were HIV+ (median CD4 count, 202 cells/mm3), 40 (10%) were pharmacologically immunosuppressed (nontransplant), and 1 had a primary immunodeficiency (Figure 1). Overall, EBV DNA was detected in the plasma and/or PBMCs in the absence of EBV+ disease in 40% of HIV+ patients (median CD4 count, 123 cells/mm3), 25% of transplant recipients, 12% of nontransplant patients on pharmacologic immunosuppression, 4% of patients with primary immunodeficiencies, and 12% of hospitalized patients with no apparent immunocompromise, with most of the positive specimens reflecting the detection of EBV DNA in PBMCs only (Table 1, with detection rates in plasma and PBMCs shown separately).
In plasma, there was no difference in EBV DNA copy number between HIV+ patients, transplant recipients, nontransplant patients on pharmacologic immunosuppression, patients with primary immunodeficiencies, and immunocompetent patients (Figure 4A). By contrast, PBMC EBV DNA copy number was higher among HIV+ patients compared with each of the other groups and also higher among transplant recipients as compared with nontransplant patients on pharmacologic immunosuppression, immunocompetent patients, and those with primary immunodeficiencies; Kruskal–Wallis P < .0001 (Figure 4B). Among those with HIV, EBV DNA copy number in PBMCs did not differ by CD4 count, although a higher HIV viral load was associated with a lower EBV DNA copy number in PBMCs (F[1,88] = 7.23; P < .01; R2 = 0.076). Among transplant recipients, EBV DNA copy number in PBMCs was not related to the organ transplanted, although immunosuppressive regimens containing steroids were associated with a higher likelihood of detecting EBV DNA in PBMCs after adjusting for the use of mycophenolate mofetil and other immunosuppressant drugs (likelihood ratio χ2 = 20.06, P = .002).
Box-and-whisker plots of EBV DNA copy number by immune status among patients with no EBV+ disease. (A) EBV DNA copy number in plasma. (B) EBV DNA copy number in PBMCs. Significant differences between groups are shown with the corresponding P values.
Box-and-whisker plots of EBV DNA copy number by immune status among patients with no EBV+ disease. (A) EBV DNA copy number in plasma. (B) EBV DNA copy number in PBMCs. Significant differences between groups are shown with the corresponding P values.
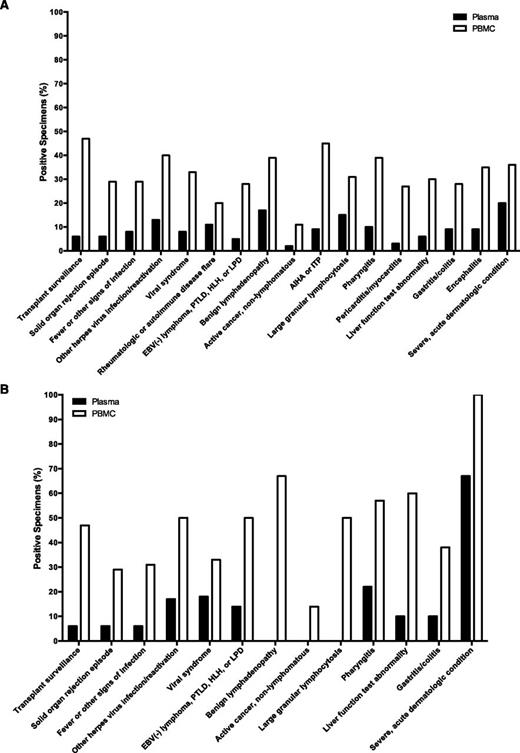
EBV DNA testing was often performed as part of diagnostic evaluations for clinical findings such as fever, hepatitis, cytopenias, lymphadenopathy, or viral syndromes. Across a wide range of indications for testing, EBV DNA was more frequently present in PBMCs than in plasma in the context of no EBV+ disease (Figure 5A). This pattern was similarly seen among transplant recipients (Figure 5B). Supplemental Table 1 shows the number of patients evaluated for EBV DNA in the blood across a range of diagnoses or reasons for testing, with the percentage of positive specimens in plasma and PBMCs for each subgroup. The EBV DNA copy numbers in plasma and PBMCs among patients with no EBV+ disease are shown in supplemental Figure 3, separated by the indication for EBV DNA testing. Non-EBV+ diagnoses associated with the highest percentages of patients with EBV DNA detected in PBMCs included routine transplant surveillance (47%), autoimmune hemolytic anemia or thrombocytopenia (45%), and reactivation of another herpes virus (40%). Clinical situations where no EBV+ diagnosis was made, but EBV DNA detection in plasma was relatively common included severe acute dermatologic conditions, such as full-body drug eruptions (20%), benign lymphadenopathy (17%), and large granular lymphocytosis (15%).
Percentage of plasma and PBMC specimens with detectable EBV DNA among patients with no EBV+ disease, grouped by the clinical setting in which EBV DNA testing was performed. (A) Patients with no EBV+ disease (n = 1419). (B) Transplant patients with no EBV+ disease (n = 562). AIHA, autoimmune hemolytic anemia; ITP, immune thrombocytopenic purpura; LPD, lymphoproliferative disorder.
Percentage of plasma and PBMC specimens with detectable EBV DNA among patients with no EBV+ disease, grouped by the clinical setting in which EBV DNA testing was performed. (A) Patients with no EBV+ disease (n = 1419). (B) Transplant patients with no EBV+ disease (n = 562). AIHA, autoimmune hemolytic anemia; ITP, immune thrombocytopenic purpura; LPD, lymphoproliferative disorder.
Discussion
In plasma, EBV DNA can be shed or extruded from benign, latently infected cells (namely B lymphocytes) or EBV+ tumor cells, or may be present as virion DNA in the setting of lytic, replicative infection. In most settings, lytic replication is not the predominant source of cell-free viral DNA in those with EBV+ malignancies or lymphoproliferative disorders.28,29 In PBMCs, EBV DNA is typically harbored within latently infected B lymphocytes. In the setting of immunocompromise, patients commonly have an increase in the absolute number of latently infected B lymphocytes, as well as a higher frequency of EBV in non-B immune cells.30,31
In patients with EBV+ tumors, most EBV DNA in plasma is tumor derived.28 By contrast, the EBV DNA copy number in the PBMC fraction likely represents very few, if any, circulating EBV+ tumor cells among those with EBV+ malignancies. In the present study, this was most clearly demonstrated in patients with EBV+ NPC, a tumor which is not of B-lymphoid origin, all of whom had EBV DNA detected in plasma but none of whom had EBV DNA detected in PBMCs, consistent with prior studies.32 In EBV+ diseases where the proliferation of latently infected, circulating cells is central to the disease process, such as IM and early EBV+ PTLD, EBV DNA copy number in PBMCs may be a reasonable marker of disease or elevated disease risk, as in the case of EBV+ PTLD. However, EBV DNA in PBMCs would not be expected to be a good marker of many EBV+ diseases, where the involved cells are not circulating intact within the blood to any significant degree. In this study, EBV DNA was commonly detected in PBMCs among those without EBV+ diseases, whereas EBV DNA was less frequently detected in plasma in the absence of an EBV+ disease and was present in plasma in all but 1 patient with an active, systemic EBV+ disease. Although a limitation of the study is the exclusion of a large number of patients with positive EBV DNA qPCRs due to insufficient follow-up, the performance of plasma remained superior to PBMCs when cases of positive EBV DNA qPCRs with shorter follow-up were included. Across a range of diseases, the current study demonstrates that cell-free (plasma) EBV DNA performs better than cellular (PBMC) EBV DNA as a marker of EBV+ diseases.
Within transplant cohorts, previous reports support the non-inferiority of EBV DNA quantification in plasma vs PBMCs as a tool for screening for EBV+ PTLD and the merits of using plasma specimens to assess treatment response.2-4,10,33,34 Although EBV DNA copy number in PBMCs will often rise in advance of EBV+ PTLD diagnosis, plasma EBV DNA appears to perform as well as PBMC specimens as an early indicator of EBV+ PTLD, with fewer false-positive tests. Importantly, fluctuations of EBV DNA copy number in plasma during therapy for EBV+ PTLD have been shown to correspond to clinical response, whereas EBV DNA copy number in PBMCs can remain elevated in times of remission or be undetectable in patients with tumor progression.10,11 Our findings further demonstrate plasma EBV DNA is a good marker of EBV+ PTLD, distinguishing those with untreated EBV+ disease from those with EBV− disease or EBV+ disease in remission.
Many clinical laboratories quantify EBV DNA in whole blood only and some transplant centers argue that whole blood is more sensitive for monitoring for EBV+ PTLD.6 A major disadvantage of whole blood, however, is the inability to discern how much EBV DNA is present in PBMCs and how much is present in plasma. Because this distinction has clinical relevance, whole blood assays may be enhanced by the additional quantification of EBV DNA in plasma in situations where the copy number in whole blood is quite elevated. There are increasing data to support the use of cell-free EBV DNA as a tumor marker. In HL, we and others have previously shown that EBV DNA is detectable in the plasma of patients untreated or relapsed EBV+ HL, but not detectable in most patients with EBV− HL or EBV+ HL in remission.21,35,36 This has also been demonstrated in other lymphomas, including those arising in immunocompromised patients,27 as well as HLH.24,37-43 In the present study, patients with active EBV+ lymphoma all had EBV DNA detected in plasma, whereas those in remission most often had no EBV DNA detected in plasma, supporting its clinical utility as a marker of disease status.
In conclusion, within a large cohort of predominantly hospitalized patients, quantification of EBV DNA in the plasma and PBMCs can serve as a marker of EBV+ disease across a range of diagnoses. Given that the detection of EBV DNA in PBMCs is not uncommon in patients without an EBV+ diagnosis, particularly among patients who are immunocompromised, plasma appears to be a better specimen source when evaluating for EBV+ disease or tracking EBV+ disease response to therapy.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported by grants from the National Cancer Institute, National Institutes of Health (R21 CA188824 and P30 CA006973) (R.F.A.) and a faculty development award to J.A.K., allocated through the Centers for AIDS Research (National Institute of Allergy and Infectious Diseases, National Institutes of Health, P30 AI094189).
Authorship
Contribution: J.A.K. collected and analyzed the data, conceptualized the clinical focus of the study, and wrote the manuscript; A.M.H. collected data; C.M.D. participated in the study concept and design, and preliminary data collection and analysis; A.B.M. performed statistical analyses; A.E.G. collected data; R.F.A. and A.V. participated in study concept and design; and A.V. directed the validation of EBV qPCR test performance characteristics, oversaw the clinical testing of patient specimens, and conceptualized and led the study. All authors participated in the editing of the manuscript and approved the final version.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for J.A.K. is National Institutes of Health, Bethesda, MD.
Correspondence: Jennifer A. Kanakry, National Institutes of Health, 10 Center Dr, Building 10/CRC, Room 4-3132, Bethesda, MD 20892; e-mail: jennifer.kanakry@nih.gov; and Alexandra Valsamakis, Johns Hopkins Hospital, 600 N. Wolfe St, Meyer Building, Room B1-193, Baltimore, MD 21287; e-mail: avalsam1@jhmi.edu.