Key Points
AP-1 complexes of the Jun/ATF type promote growth of ABC DLBCL cell lines.
High expression of ATF3 is a hallmark of samples from patients with non-GC/ABC DLBCL.
Abstract
A hallmark of the diffuse large B-cell lymphoma (DLBCL) of the activated B-cell (ABC) type, a molecular subtype characterized by adverse outcome, is constitutive activation of the transcription factor nuclear factor–κB (NF-κB), which controls expression of genes promoting cellular survival and proliferation. Much less, however, is known about the role of the transcription factor activator protein-1 (AP-1) in ABC DLBCL. Here, we show that AP-1, like NF-κB, was controlled by constitutive activation of the B-cell receptor signaling component caspase recruitment domain-containing membrane-associated guanylate kinase 1 (CARMA1) and/or the Toll-like receptor signaling component myeloid differentiation primary response gene 88 (MyD88) in ABC DLBCL cell lines. In contrast to germinal center (GC) B-cell (GCB) DLBCL, ABC DLBCL cell lines expressed high levels of the AP-1 family members c-Jun, JunB, and JunD, which formed heterodimeric complexes with the AP-1 family members activating transcription factor (ATF) 2, ATF3, and ATF7. Inhibition of these complexes by a dominant-negative approach led to impaired growth of a majority of ABC DLBCL cell lines. Individual silencing of c-Jun, ATF2, or ATF3 decreased cellular survival and revealed c-Jun/ATF2-dependent control of ATF3 expression. As a consequence, ATF3 expression was much higher in ABC vs GCB DLBCL cell lines. Samples derived from DLBCL patients showed a clear trend toward high and nuclear ATF3 expression in nodal DLBCL of the non-GC or ABC subtype. These findings identify the activation of AP-1 complexes of the Jun/ATF-type as an important element controlling the growth of ABC DLBCL.
Introduction
Diffuse large B-cell lymphoma (DLCBL) is the most frequent form of lymphoid cancer, accounting for 30% to 35% of all nodal lymphomas.1 Based on gene expression profiling (GEP), 3 distinct subtypes of DLBCL have been identified, namely the germinal center (GC) B-cell (GCB), activated B-cell (ABC), and primary mediastinal B-cell lymphoma subtypes.2 The ABC subtype of DLBCL is characterized by adverse prognosis and constitutive activation of the transcription factor nuclear factor–κB (NF-κB).3 This is thought to be the consequence of somatic mutations in the genes encoding the B-cell receptor (BCR)-associated CD79A and CD79B chains,4 or the BCR signal transducer caspase recruitment domain-containing membrane-associated guanylate kinase-1 (CARMA1) (also known as CARD11),5 and polymorphisms in RNF31 (also known as HOIP),6 which result in constitutive BCR signaling. These can be present alone or in combination with activating mutations in genes encoding the Toll-like receptor (TLR) downstream signaling protein MyD887 and inactivation and/or deletion of the gene encoding A20, a negative regulator of the NF-κB pathway.8 As a consequence of these mutations, ABC DLBCL have a constitutive activation of NF-κB via BCR- and/or TLR-signaling pathways, whose natural physiological role is to promote B-cell proliferation and survival.1
Natural engagement of the antigen receptor, or of TLRs, activates not only NF-κB, but also transcription factors of the activator protein-1 (AP-1) family.9,10 However, little is known about the relevance of the AP-1 transcription factor family and the molecular pathways triggering its activation leading to pathology of ABC DLBCL. The AP-1 family comprises hetero- and homodimeric transcription factors that are formed by combinations of members of the Jun, Fos, activating transcription factor (ATF), and Maf subfamilies.11 AP-1 dimers have different DNA recognition sequences and are also differentially regulated according to the cell type and/or activating stimulus. Various posttranslational modifications can modulate AP-1 activity, by controlling the abundance and the activity of the individual dimers.12 Ser/Thr kinases of the mitogen-activated protein kinase (MAPK) family, in particular, extracellular signal-regulated kinase (ERK) and c-Jun N-terminal kinase (JNK), have been shown to phosphorylate c-Fos and c-Jun and to thereby control their stability and activity.11 Antigen receptor triggering leads to induction and/or activation of multiple AP-1 family members, including c-Fos, c-Jun, JunB, ATF2, and ATF3, but the individual roles of these AP-1 family transcription factors for lymphocyte proliferation remain poorly understood.11,13-17
Recent studies have provided some insight into how the BCR and the TLR signaling pathways activate gene transcription via the AP-1 pathway.18 BCR signaling depends on the CARMA1-BCL10-MALT1 (CBM) complex, whereas TLR signaling depends on the adaptor protein MyD88 and the kinases interleukin-1 receptor-associated kinase (IRAK) 1 and IRAK4. The ubiquitin ligase tumor necrosis factor receptor-associated factor 6 and the Ser/Thr kinase transforming growth factor beta-activated kinase 1 (TAK1) are downstream targets of both BCR/CBM- and TLR/MyD88/IRAK-dependent signals. TAK1 has been reported to act as an upstream regulator of JNK19-22 ; it is thus generally assumed that BCR- and TLR-induced JNK activation is crucial for AP-1 activation. However, the exact role of JNK in the activation of individual AP-1 family heterodimers in lymphocytes remains incompletely understood, and whether JNK is strictly required for AP-1 activation in lymphocytes remains controversial.23-26 Another particular difficulty in studying the role of AP-1 transcription factors is the fact that the AP-1 family comprises >20 members that can form numerous different heterodimers with partially redundant functions and highly diverse mechanisms of regulation.11 Therefore, little is currently known about the exact composition and relevance of AP-1 complexes in activated lymphocytes and the development of ABC DLBCL.
Here, we show that cell lines derived from ABC DLBCL are characterized by constitutive upregulation of c-Jun, JunB, and ATF3, which was mediated by CARMA1 and MyD88. Jun members formed complexes with ATF2 or ATF7 (complexes of type I), or with ATF3 (complexes of type II). Inhibition of these complexes by a dominant-negative approach impaired the viability of most ABC DLBCL cell lines. Among the different members of the complexes, ATF3, ATF2, and c-Jun were the main drivers of cellular survival. Interestingly, ATF3, but not ATF2 or ATF7, was exclusively expressed in cell lines derived from the ABC subtype of DLBCL, and immunohistochemical analysis of biopsies of DLBCL patients confirmed preferential strong and nuclear ATF3 staining in samples from patients with nodal lymphoma of the non-GC or ABC subtype of DLBCL. Collectively, these findings identify the activation of specific AP-1 complexes as a hallmark of ABC DLBCL.
Methods
Cellular transfection and transduction
Lentiviral transduction and viability assays have been described.27 Retroviral transduction was achieved with A-Fos subcloning into the retroviral vector pMSCV-IRES-GFP.28 Expression of A-Fos in DLBCL lines was achieved by retroviral transduction as described previously.29 The transduced cells were monitored for live GFP+ cells by flow cytometry as described.30
Cell culture, cell stimulation, and reporter assays
Jurkat cells and DLBCL cell lines BJAB, SUDHL-4, SUDHL-6, HT, HBL-1, OCI-Ly3, OCI-Ly10, and TMD8 were cultured as described.30 For stimulation of Jurkat T cells, a mixture of phorbol 12-myristate 13-acetate (PMA; 10 ng/mL; Alexis) and ionomycin (1 μM; Calbiochem) was used. In some experiments, cells were preincubated with 1 μM of the JNK inhibitor SP600125 (Calbiochem), 1 μM of the TAK1 inhibitor 5Z7 (Sigma-Aldrich), protein kinase C (PKC) inhibitors (500 nM bisindolylmaleimide VIII acetate [Alexis] or 1 μM Gö6976 [Calbiochem]), or with corresponding volumes of solvent for the indicated times at 37°C. Interleukin-2 (IL-2) luciferase assays were performed as described.28
Cell lysis, immunoprecipitation, and western blot analysis
Cells were lysed in radioimmunoprecipitation assay buffer containing 50 mM Tris-HCl, pH 7.4, 1% Nonidet-P40, 0.25% sodium deoxycholate, 150 mM NaCl, and 1 mM EDTA, or in lysis buffer containing 50 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid), pH 7.5, 150 mM NaCl, 1% Triton X-100, protease inhibitors (Complete; Roche), and phosphatase inhibitors (NaF, Na4P2O7, and Na3VO4). In some experiments, lysates were incubated with or without 0.5 mM BS3 (Thermo Scientific) for 1 hour at 4°C. Immunoprecipitation, sample analysis by high-resolution sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), and immunoblot were performed as described.28
Patient populations
The construction of the tissue microarray (TMA) with samples from de novo previously untreated DLBCL patients classified by the Hans algorithm31 has been described elsewhere.32 Of the original cohort of 109 patients, sufficient tumor tissue for additional immunohistochemical analyses was available from 70 patients. The local ethics committee approved this retrospective analysis (KEK 160/14). A second cohort of DLBCL patients consisted of a GEP-characterized subset of a patient cohort already published.29 The study of this cohort was approved by the ethics committee of Northwestern and Central Switzerland (EKNZ 2014-252).
Immunohistochemistry for ATF3
Immunohistochemical staining of the ATF3 protein was performed on the TMA slides using an automated immunostainer (Leica BOND-III; Leica Biosystems). As a pretreatment for antigen retrieval, the TMA section was placed in epitope retrieval solution (Tris buffer for 30 minutes at 95°C). Subsequently, the TMA was incubated at room temperature with rabbit anti-human ATF3 antibody (sc-188; Santa Cruz Biotechnology) at a working concentration of 1:50 for 30 minutes. Antigen detection was performed using a commercial detection kit (Bond Polymer Refine Detection) with diaminobenzidin as the chromogen.
Immunohistochemical scoring
For assessment of ATF3 expression, the stained TMA slides were scanned with Aperio (Vista) and evaluated at ×40 magnification. The staining intensity was stratified from 0 to 2, where 0 (designated “weak”) = negative or weak staining in <10% of neoplastic cells, 1 (designated “intermediate”) = weak or moderate staining in >10% neoplastic cells, and 2 (designated “strong”) = strong staining in >10% of neoplastic cells. Staining was further categorized by its pattern as partially or predominantly nuclear or cytoplasmic. Staining intensity was semiquantitative and used internal samples as reference points.
Plasmids, antibodies, mass spectrometry, and fluorescence microscopy
Detailed information about plasmids, antibodies, mass spectrometry, and fluorescence microscopy can be found in the supplemental Methods (available on the Blood Web site).
Statistical analysis
The 2-tailed Student t test was used for statistical analysis; values of P ≤ .05 were considered statistically significant.
Results
Jun family proteins are upregulated in ABC DLBCL cell lines in a CARMA1/MALT1- and MyD88/IRAK-dependent manner
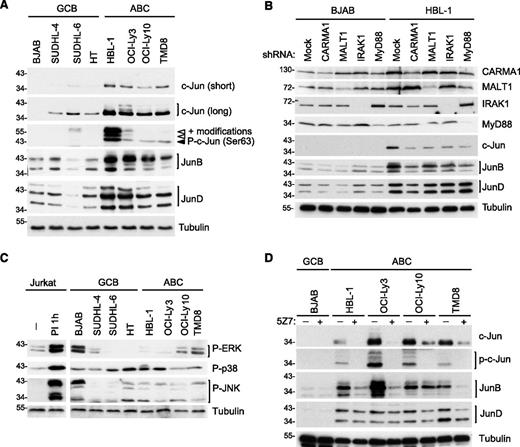
To assess whether AP-1 family members are differentially expressed in ABC vs GCB DLBCL, we first monitored the expression of different Jun family members in 4 cell lines derived from each of the 2 DLBCL subtypes. Interestingly, c-Jun and JunB protein levels were clearly higher in all ABC DLBCL cell lines compared with GCB DLBCL cell lines (Figure 1A), consistent with a recent report.18 In addition, JunD levels were generally higher in ABC DLBCL cell lines (Figure 1A). Most of the cell lines derived from ABC DLBCL, including all 4 cell lines used in this study, have somatic mutations driving constitutive BCR/CBM- or TLR/MyD88-dependent signaling.4,5,7,8,33 We thus subsequently assessed the individual requirement of these pathways for the expression of Jun family members. Expression of c-Jun and JunB, but not of JunD, was clearly dependent on constitutive CBM- and MyD88-dependent constitutive signaling, as evident from the observed reduction of c-Jun and JunB expression upon silencing of CARMA1, MALT1, MyD88, or IRAK1 (Figure 1B). Consistent with a critical role of PKC family kinases downstream of CD79 and upstream of CARMA1,34-36 we observed a reduction of cellular c-Jun protein expression in all ABC DLBCL cell lines with CD79 mutations (HBL-1, OCI-Ly10, and TMD8) upon pretreatment with the pan-PKC inhibitor bisindolylmaleimide VIII (BIM VIII) or the more selective inhibitor of classical PKC isoforms, Gö6976, with the exception of the HBL-1 cells, which did not react to Gö6976 (supplemental Figure 1A).
Upregulation of c-Jun and JunB in ABC DLBCL cell lines is CARMA1-, MALT1-, MyD88-, IRAK1-, and TAK1-dependent. (A) Analysis of c-Jun, JunB, and JunD protein expression and c-Jun phosphorylation on Ser 63 in GCB and ABC cell lines by western blot. (B) Analysis of c-Jun, JunB, and JunD protein expression in lysates of HBL-1 (ABC) and BJAB (GCB) cell lines transduced with control small hairpin RNA (shRNA) or with CARMA1-, MALT1-, IRAK1-, or Myd88-specific shRNA. Silencing efficiency was assessed by western blot analysis using anti-CARMA1, anti-MALT1, anti-IRAK1, and anti-MyD88 antibodies. (C-D) Protein expression in GCB and ABC DLBCL cell lines was determined by western blot using the indicated antibodies. In panel C, we used lysates of Jurkat cells treated with PMA and ionomycin (PI) for 1 hour as a positive control for MAPK activation. In panel D, DLBCL cell lines of the GCB (BJAB), or ABC subtype (all others) were treated with the TAK1 inhibitor 5Z7 or with solvent alone for 24 hours. In all figure panels, blotting for tubulin served as a loading control. Data are representative of at least 3 (A-B) or 2 (C-D) independent experiments.
Upregulation of c-Jun and JunB in ABC DLBCL cell lines is CARMA1-, MALT1-, MyD88-, IRAK1-, and TAK1-dependent. (A) Analysis of c-Jun, JunB, and JunD protein expression and c-Jun phosphorylation on Ser 63 in GCB and ABC cell lines by western blot. (B) Analysis of c-Jun, JunB, and JunD protein expression in lysates of HBL-1 (ABC) and BJAB (GCB) cell lines transduced with control small hairpin RNA (shRNA) or with CARMA1-, MALT1-, IRAK1-, or Myd88-specific shRNA. Silencing efficiency was assessed by western blot analysis using anti-CARMA1, anti-MALT1, anti-IRAK1, and anti-MyD88 antibodies. (C-D) Protein expression in GCB and ABC DLBCL cell lines was determined by western blot using the indicated antibodies. In panel C, we used lysates of Jurkat cells treated with PMA and ionomycin (PI) for 1 hour as a positive control for MAPK activation. In panel D, DLBCL cell lines of the GCB (BJAB), or ABC subtype (all others) were treated with the TAK1 inhibitor 5Z7 or with solvent alone for 24 hours. In all figure panels, blotting for tubulin served as a loading control. Data are representative of at least 3 (A-B) or 2 (C-D) independent experiments.
c-Jun and JunB expression requires TAK1 activity
The exact molecular mechanism that controls JunB and JunD upregulation in lymphocytes is unknown. c-Jun, however, is stabilized by phosphorylation on Ser residues 63 and 73, which inhibits its otherwise constitutive proteasomal degradation.37,38 Accordingly, the increased levels of c-Jun expression in ABC DLBCL cell lines correlated with constitutive c-Jun phosphorylation on Ser 63 (Figure 1A). Because phosphorylation on Ser 63 has been described to be a target of phosphorylation by the MAPK JNK,37,38 we analyzed the activation status of the MAPK JNK, as well as other MAPKs such as p38 and ERK. We detected constitutive JNK, p38, and ERK activation in several ABC and GCB DLBCL cell lines (Figure 1C), in agreement with recently reported findings.39 However, we saw no obvious correlation between MAPK activation and c-Jun phosphorylation and accumulation (compare Figure 1, panels A and C). When pretreating the DLBCL cell lines with the JNK inhibitor SP600125, at a concentration that efficiently prevented PMA/ionomycin-induced c-Jun accumulation in Jurkat T cells (supplemental Figure 1B), we observed a clear reduction of c-Jun levels only in the ABC DLBCL cell line HBL-1 (supplemental Figure 1C). Thus, JNK activation is unlikely to be a common driving factor of c-Jun phosphorylation on Ser 63, which is a specific feature of ABC DLBCL cell lines. In contrast, pretreatment of the cells with an inhibitor of the Ser/Thr kinase TAK1, 5Z7, which has been reported to act as a downstream signaling component of antigen receptor and TLR-induced signaling and an upstream regulator of JNK,19-22 efficiently reduced c-Jun expression levels and phosphorylation in all ABC DLBCL cell lines tested (Figure 1D). The TAK1 inhibitor also affected JunB levels in 3 of 4 ABC cell lines, whereas it had little, or no effect, on JunD levels (Figure 1D). Thus, cell lines derived from ABC DLBCL have high levels of JunB and c-Jun expression and c-Jun phosphorylation. Whether this is mediated directly or indirectly by TAK1 and/or a TAK1-dependent kinase remains to be identified.
Jun family members form constitutive type I complexes with ATF2 or ATF7 in ABC and GCB DLBCL cell lines
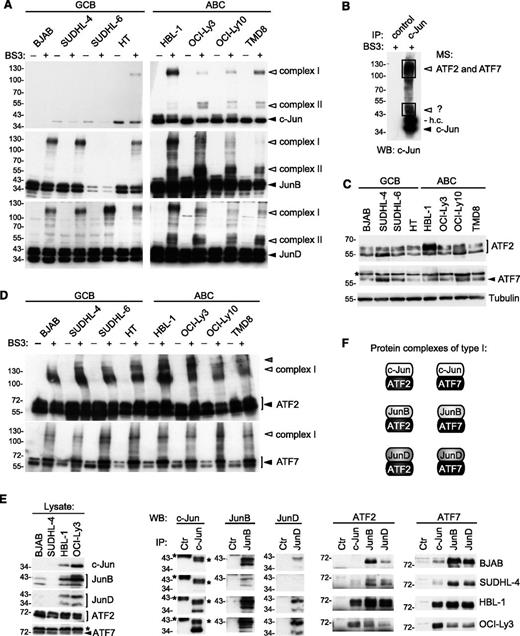
AP-1 family members can form homo- or heterodimeric complexes, typically by association of 2 AP-1 family members of the Jun, Fos, ATF, and Maf subfamilies.11 To gain additional insight into the biochemical composition of AP-1 complexes formed by c-Jun, JunB, and JunD in ABC DLBCL cell lines, we performed chemical cross-linking experiments. Upon treatment of cell lysates with the chemical cross-linker BS3, we observed that all 3 Jun family members formed 2 different types of higher molecular-weight complexes, subsequently called complexes of type I or II (Figure 2A). The observed distinct shifts in the relative molecular weight of the Jun-binding complexes suggested that these contained Jun-binding partners of 2 distinct molecular weights. For c-Jun, type I complexes tended to be more abundant in ABC than in GCB DLBCL cell lines, whereas formation of type II complexes was a distinct feature of all Jun family members specifically observed in ABC, but not in GCB, DLBCL cell lines. By mass spectrometry, we identified ATF2 and ATF7 as specific components of type I complexes of c-Jun in HBL-1 cells (Figure 2B; supplemental Table 1), suggesting that type I complexes contain both c-Jun/ATF2 and c-Jun/ATF7 heterodimers. ATF2 and ATF7 expression levels were similar in all ABC and GCB cell lines tested, with the exception of HBL-1 cells, which had slightly higher ATF2 expression levels (Figure 2C). Cross-linking of lysates using BS3 revealed that ATF2 and ATF7 formed high-molecular-weight complexes in both GCB and ABC DLBCL cell lines, and that these complexes corresponded in molecular weight to type I complexes (Figure 2D). Association of ATF2 and ATF7 with c-Jun, JunB, and JunD was confirmed by coimmunoprecipitation assays performed on 2 ABC and 2 GCB DLBCL cell lines (Figure 2E). Among these, c-Jun complexes with ATF2 and ATF7 were more abundant in ABC than in GCB DLBCL cell lines (Figure 2E). Thus, the Jun family members c-Jun, JunB, and JunD form constitutive complexes with ATF2 or ATF7 (type I complexes; Figure 2F) in both ABC and GCB DLBCL cell lines. However, c-Jun-containing type I complexes were more abundant in ABC DLBCL cell lines (Figure 2A,E).
Jun subunits form heterodimers with ATF2 or ATF7 in ABC DLBCL cell lines. (A) Lysates from the indicated ABC and GCB DLBCL cell lines were treated with the cross-linker BS3 or with solvent alone for 1 hour at 4°C. Cross-linked (open arrowheads) and non–cross-linked (filled arrowheads) proteins were revealed by western blot using anti-c-Jun, anti-JunB, and anti-JunD antibodies. (B) BS3-treated HBL-1 cell lysates were immunoprecipitated with anti-c-Jun beads or beads alone, separated by SDS-PAGE and stained with Coomassie. Proteins present in protein complexes of type I and II were analyzed by mass spectrometry (MS). ATF2 and ATF7 were identified to be part of the protein complex type I, whereas not enough material was present in protein complex type II for MS identification. h.c., heavy chain of c-Jun. (C) Protein expression in GCB and ABC cell lines was determined by western blot using anti-ATF2 and anti-ATF7 antibodies. (D) As in panel A, but proteins were revealed using anti-ATF2 and anti-ATF7 antibodies. Gray arrowhead indicates a third, unidentified type of complex. (E) Two ABC and 2 GCB cell lines were lysed and proteins were precipitated using anti-c-Jun, anti-JunB, anti-JunD, or anti-IRE1α antibodies (Ctr, control antibody). Proteins in lysates were analyzed by western blotting with anti-c-Jun, anti-JunB, and anti-JunD antibodies, and coprecipitating proteins (IP) with anti-ATF2 and anti-ATF7 antibodies, as indicated. (F) The composition of protein complexes of type I is schematically depicted. Data are representative of 3 (A,C) and 2 (D,E) independent experiments. *A nonspecific band recognized by anti-ATF7 (C,E) and migration of antibody heavy chains (E). WB, western blot.
Jun subunits form heterodimers with ATF2 or ATF7 in ABC DLBCL cell lines. (A) Lysates from the indicated ABC and GCB DLBCL cell lines were treated with the cross-linker BS3 or with solvent alone for 1 hour at 4°C. Cross-linked (open arrowheads) and non–cross-linked (filled arrowheads) proteins were revealed by western blot using anti-c-Jun, anti-JunB, and anti-JunD antibodies. (B) BS3-treated HBL-1 cell lysates were immunoprecipitated with anti-c-Jun beads or beads alone, separated by SDS-PAGE and stained with Coomassie. Proteins present in protein complexes of type I and II were analyzed by mass spectrometry (MS). ATF2 and ATF7 were identified to be part of the protein complex type I, whereas not enough material was present in protein complex type II for MS identification. h.c., heavy chain of c-Jun. (C) Protein expression in GCB and ABC cell lines was determined by western blot using anti-ATF2 and anti-ATF7 antibodies. (D) As in panel A, but proteins were revealed using anti-ATF2 and anti-ATF7 antibodies. Gray arrowhead indicates a third, unidentified type of complex. (E) Two ABC and 2 GCB cell lines were lysed and proteins were precipitated using anti-c-Jun, anti-JunB, anti-JunD, or anti-IRE1α antibodies (Ctr, control antibody). Proteins in lysates were analyzed by western blotting with anti-c-Jun, anti-JunB, and anti-JunD antibodies, and coprecipitating proteins (IP) with anti-ATF2 and anti-ATF7 antibodies, as indicated. (F) The composition of protein complexes of type I is schematically depicted. Data are representative of 3 (A,C) and 2 (D,E) independent experiments. *A nonspecific band recognized by anti-ATF7 (C,E) and migration of antibody heavy chains (E). WB, western blot.
ATF3 is specifically upregulated and forms type II complexes with Jun family members in ABC DLBCL cell lines
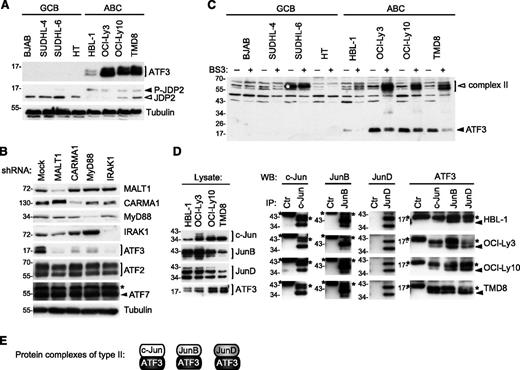
Protein components present in type II complexes were not sufficiently abundant to be identified by mass spectrometry. However, based on the relative molecular weight of the chemically cross-linked type II complexes, we hypothesized that these may correspond to lower molecular-weight members of the ATF subfamily, such as ATF3 and JDP2. In line with this idea, we found that ATF3 was specifically expressed in cell lines derived from ABC, but not from GCB, DLBCLs (Figure 3A). In contrast, the unconventional ATF family member JDP2, which acts as a repressor of ATF2 and ATF3 function,40,41 was less abundant in 3 of 4 ABC as compared with GCB DLBCL cell lines. JDP2 has been shown to be targeted for proteasomal degradation upon its phosphorylation.42 Indeed, all ABC DLBCL cell lines were characterized by the presence of an additional slower-migrating, phosphatase-sensitive isoform of JDP2 (Figure 3A; data not shown), suggesting that JDP2 is constitutively phosphorylated and targeted for proteasomal degradation in ABC DLBCL cell lines.
ATF3 is overexpressed and forms heterodimers with Jun subunits in ABC DLBCL cell lines. (A) Analysis of ATF3 and JDP2 protein expression in GCB and ABC cell lines was determined by western blot using ATF3 and JDP2 antibodies. Filled arrowhead indicates the position of phosphorylated JDP2; open arrowheads indicate nonphosphorylated JDP2. (B) Immunoblot analysis of lysates of HBL-1 (ABC DLBCL) and BJAB (GCB DLBCL) cell lines, transduced with control shRNA or with CARMA1-, MALT1-, IRAK1-, or MyD88-specific shRNA. Silencing efficiency was assessed by western blot analysis using anti-CARMA1, anti-MALT1, anti-IRAK1, and anti-MyD88 antibodies. ATF2, ATF3, ATF7 protein levels were assessed by western blot. Blotting for tubulin served as loading control in panels A and B. (C) Lysates of indicated ABC and GCB cell lines were treated with the cross-linker BS3 or with solvent alone for 1 hour at 4°C. Cross-linked (open arrowheads) and non–cross-linked (filled arrowheads) proteins were assessed by western blot using anti-ATF3 antibodies. White asterisk indicates the position of a nonspecific band detected by anti-ATF3. (D) ABC DLBCL cell lines were immunoprecipitated with anti-c-Jun, anti-JunB, anti-JunD, or anti-IRE1α antibodies (Ctr, control antibody). Immunoprecipitated proteins (IPs) were assessed with anti-c-Jun, anti-JunB, and anti-JunD antibodies, and coprecipitating proteins (co-IP) detected by anti-ATF3. *Heavy chain or light chain of the c-Jun, JunB, JunD, and control antibodies; filled arrowhead indicates ATF3 in the co-IP. (E) The composition of protein complexes of ATF3 with Jun family members (type 2 complexes) is depicted. Data are representative of 3 (A) and 2 (B-D) independent experiments.
ATF3 is overexpressed and forms heterodimers with Jun subunits in ABC DLBCL cell lines. (A) Analysis of ATF3 and JDP2 protein expression in GCB and ABC cell lines was determined by western blot using ATF3 and JDP2 antibodies. Filled arrowhead indicates the position of phosphorylated JDP2; open arrowheads indicate nonphosphorylated JDP2. (B) Immunoblot analysis of lysates of HBL-1 (ABC DLBCL) and BJAB (GCB DLBCL) cell lines, transduced with control shRNA or with CARMA1-, MALT1-, IRAK1-, or MyD88-specific shRNA. Silencing efficiency was assessed by western blot analysis using anti-CARMA1, anti-MALT1, anti-IRAK1, and anti-MyD88 antibodies. ATF2, ATF3, ATF7 protein levels were assessed by western blot. Blotting for tubulin served as loading control in panels A and B. (C) Lysates of indicated ABC and GCB cell lines were treated with the cross-linker BS3 or with solvent alone for 1 hour at 4°C. Cross-linked (open arrowheads) and non–cross-linked (filled arrowheads) proteins were assessed by western blot using anti-ATF3 antibodies. White asterisk indicates the position of a nonspecific band detected by anti-ATF3. (D) ABC DLBCL cell lines were immunoprecipitated with anti-c-Jun, anti-JunB, anti-JunD, or anti-IRE1α antibodies (Ctr, control antibody). Immunoprecipitated proteins (IPs) were assessed with anti-c-Jun, anti-JunB, and anti-JunD antibodies, and coprecipitating proteins (co-IP) detected by anti-ATF3. *Heavy chain or light chain of the c-Jun, JunB, JunD, and control antibodies; filled arrowhead indicates ATF3 in the co-IP. (E) The composition of protein complexes of ATF3 with Jun family members (type 2 complexes) is depicted. Data are representative of 3 (A) and 2 (B-D) independent experiments.
As for c-Jun and JunB, the expression of ATF3 was strongly reduced upon silencing of CARMA1, MALT1, IRAK1, and MyD88 (Figure 3B). Upon treatment with the chemical cross-linker BS3, ATF3 shifted toward a higher molecular-weight complex that corresponded in size to the previously described type II complexes (Figure 3C). Moreover, ATF3 could easily be coimmunoprecipitated with c-Jun, JunB, and JunD in lysates of ABC DLBCL cell lines (Figure 3D). Collectively, these findings suggest that the AP-1 family member ATF3 is selectively expressed in cell lines derived from ABC DLBCL, and forms constitutive complexes with c-Jun, JunB, and JunD (type II complexes; Figure 3E) in these cells.
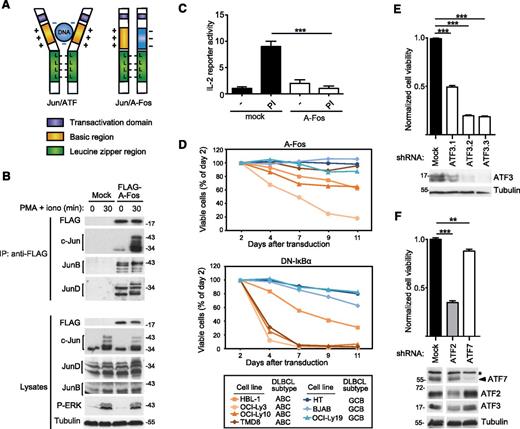
Inhibition of Jun family members by A-Fos impairs the viability of cell lines derived from ABC DLBCL
To explore the role of Jun/ATF-type dimers in ABC DLBCL, we made use of a previously described dominant-negative A-Fos construct.43 A-Fos contains the Jun-binding leucine zipper region of c-Fos fused to a negatively charged protein domain that binds to, and thereby masks, the positively charged DNA-binding domain of c-Jun (Figure 4A). When stably expressed in Jurkat T cells, the A-Fos construct bound to c-Jun, JunB, and JunD in both unstimulated and PMA/ionomycin-stimulated cells (Figure 4B), and efficiently inhibited the inducible expression of an IL-2 luciferase reporter gene (Figure 4C), which is known to be AP-1 dependent.44 We then transduced DLBCL cell lines with a retroviral construct allowing the coexpression of A-Fos with green fluorescent protein (GFP), to specifically monitor the viability of GFP+, A-Fos expressing cells. Compared with a dominant-negative IκB (DN-IκB) construct, which inhibits the NF-κB transcriptional pathway and rapidly affects the viability of ABC DLBCL cell lines, A-Fos expression led to a slow reduction in cell viability in 3 of 4 ABC DLBCL cell lines tested (Figure 4D). No impact on cell viability was observed for GCB DLBCL cell lines transduced with either DN-IκB or A-Fos (Figure 4D). To further explore the role of individual ATF family members, we silenced the expression of ATF2, ATF3, and ATF7 in HBL-1 cells and monitored cell survival (Figure 4E-F). Under these conditions, ATF3 silencing affected cell survival to an extent that was similar to that previously seen with A-Fos (Figure 4E). Silencing of ATF7 had little effect on survival, whereas ATF2 silencing clearly affected cell survival (Figure 4F). However, silencing of ATF2 simultaneously diminished the expression of ATF3, suggesting that the effect of ATF2 silencing on cell viability may be indirectly mediated by ATF3 (Figure 4F). Collectively, this suggests that ATF3-containing type II complexes have a major role in cell survival, and that ATF2-containing type I complexes most likely contribute to cell survival indirectly by affecting ATF3 levels. Silencing of c-Jun, the major component of type I complexes in ABC DLBCL cell lines (Figure 2A,E), also had a clear impact on both cell viability and ATF3 expression in HBL-1 cells (supplemental Figure 2). Together with our biochemical data (Figure 2A,E), these findings suggest that c-Jun/ATF2-dependent ATF3 expression is relevant for the viability of a majority of ABC DLBCL cell lines.
Inhibition of AP-1 complexes impairs the viability of cell lines derived from ABC DLBCL. (A) Schematic representation of the structure of transcriptionally active Jun/ATF and inactive Jun/A-Fos complexes illustrating the dominant-negative function of A-Fos. (B) Jurkat T cells were lentivirally transduced with a FLAG-tagged expression construct for A-Fos or an empty vector (mock) as control. Cells were treated with PMA and ionomycin (PI) for the indicated times. Lysates were immunoprecipitated using anti-FLAG sepharose beads and analyzed by immunoblot with the indicated antibodies. (C) Jurkat T cells were electroporated with an IL-2 firefly luciferase reporter and a renilla luciferase reporter, stimulated with PMA and ionomycin for 14 hours and the relative luciferase activity of the cell lysates was determined. (D) Viability of ABC DLBCL and GCB DLBCL cell lines transduced with constructs coexpressing GFP with FLAG-tagged A-Fos (top panel) or DN-IκBα (bottom panel), assessed by flow cytometry. (E-F) HBL-1 cells were transduced with indicated silencing constructs and cell viability was assessed using a PMS/MTS (phenazine methosulfate/3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) assay. Efficiency of protein silencing and equal loading (tubulin) was verified by western blot. *A nonspecific band recognized by anti-ATF7. Three independent shRNAs were used for ATF3. (C,E,F) Bars represent means ± standard deviation (SD); differences were statistically significant with **P < .01, ***P < .001 (unpaired Student t test). Data in panels B-E are representative of at least 2 independent experiments.
Inhibition of AP-1 complexes impairs the viability of cell lines derived from ABC DLBCL. (A) Schematic representation of the structure of transcriptionally active Jun/ATF and inactive Jun/A-Fos complexes illustrating the dominant-negative function of A-Fos. (B) Jurkat T cells were lentivirally transduced with a FLAG-tagged expression construct for A-Fos or an empty vector (mock) as control. Cells were treated with PMA and ionomycin (PI) for the indicated times. Lysates were immunoprecipitated using anti-FLAG sepharose beads and analyzed by immunoblot with the indicated antibodies. (C) Jurkat T cells were electroporated with an IL-2 firefly luciferase reporter and a renilla luciferase reporter, stimulated with PMA and ionomycin for 14 hours and the relative luciferase activity of the cell lysates was determined. (D) Viability of ABC DLBCL and GCB DLBCL cell lines transduced with constructs coexpressing GFP with FLAG-tagged A-Fos (top panel) or DN-IκBα (bottom panel), assessed by flow cytometry. (E-F) HBL-1 cells were transduced with indicated silencing constructs and cell viability was assessed using a PMS/MTS (phenazine methosulfate/3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) assay. Efficiency of protein silencing and equal loading (tubulin) was verified by western blot. *A nonspecific band recognized by anti-ATF7. Three independent shRNAs were used for ATF3. (C,E,F) Bars represent means ± standard deviation (SD); differences were statistically significant with **P < .01, ***P < .001 (unpaired Student t test). Data in panels B-E are representative of at least 2 independent experiments.
Strong nuclear ATF3 expression characterizes nodal tumors from non-GC/ABC DLBCL patients
To validate the relevance of our cell line–based findings, we first assessed the expression of ATF3 in tissue samples from a cohort of 350 patient samples classified by GEP.45 At the messenger RNA (mRNA) level, expression of ATF3, but not of ATF2 or ATF7, was significantly higher in patients with ABC vs GCB DLBCL (Figure 5A). Subsequently, we assessed ATF3 expression in DLBCL samples by immunohistochemistry (IHC). To this purpose, staining conditions for ATF3 were optimized in DLBCL cell lines to detect strong staining in all 4 ABC DLBCL cell lines tested, and only minimal background staining in GCB DLBCL cell lines (Figure 5B). We then analyzed samples of 2 different cohorts of DLBCL patients for ATF3 expression, which was scored according to the intensity (weak, intermediate, or strong; for scoring details, see “Methods”) and the preferential localization (cytoplasmic, exclusively or partially nuclear). First, we analyzed a cohort of 70 DLBCL samples, which were classified into GCB and non-GC DLBCL according to the Hans algorithm,32 and were derived from 28 nodal and 42 extranodal tumors. These IHC analyses showed various expression patterns, which differed in intensity and staining pattern; examples of these are shown in Figure 5C. When scoring ATF3 expression intensities and patterns in the combined samples, we observed a small tendency toward higher and nuclear expression in non-GC vs GCB samples (supplemental Figure 3). This tendency was striking in samples obtained from nodal tumors (Figure 5D). In contrast, no such tendency was present in samples from extranodal tumors (data not shown), most likely because the Hans algorithm has been established and validated only for nodal DLBCL. Interestingly, in gastric DLBCL, the Hans and another algorithm failed in identifying prognostically relevant subgroups.46 Furthermore, extranodal DLBCL outside of the testicles and the central nervous system more commonly encompass morphologically and clinically unrecognizable transformed marginal zone B-cell lymphomas, for which an algorithm has not been developed. We also analyzed a second cohort of 17 nodal DLBCL samples, which were classified into the GCB or ABC subtype according to GEP.29 Among these, ATF3 expression was strong in 7 of 9 samples and intermediate in 2 of 9 ABC DLBCL, whereas GCB DLBCL showed negative or intermediate expression levels in 7 of 8 samples and strong staining was observed in only 1 GCB DLBCL sample (Figure 5E). In the majority of non-GC DLBCL samples (14 of 19), or GEP-identified ABC DLBCL samples (6 of 9), the ATF3 staining was partially or predominantly nuclear. Collectively, these data identify high and preferentially nuclear protein expression of ATF3 as a hallmark of human non-GC/ABC DLBCL.
Strong nuclear ATF3 expression is a hallmark of non-GC and ABC DLBCL patients. (A) Relative mRNA expression of ATF2, ATF3, and ATF7 in ABC vs GCB DLBCL biopsies. Error bars indicate standard error of the means (SEMs). (B) Immunofluorescence staining of ATF3 expression and localization in the indicated GCB and ABC DLBCL cell lines. Bar, 5 μm. (C) Histologic staining of ATF3 expression in representative biopsy samples of DLBCL patients. Bar, 50 μM; magnification, ×400. (D-E) TMA analysis of DLBCL biopsies previously categorized into GCB or non-GC using the Hans algorithm (D), or into ABC or GCB by GEP (E). Bar graphs summarizing the classification of non-GC and GCB patients (D) or ABC and GCB patients (E) according to staining intensity (weak, intermediate [int.], and strong) and subcellular localization of ATF3 (C, cytoplasmic; N, partially or predominantly nuclear). Analysis was performed on 28 (D) or 17 (E) patients with nodal (lymph node) tumors. DAPI, 4,6 diamidino-2-phenylindole.
Strong nuclear ATF3 expression is a hallmark of non-GC and ABC DLBCL patients. (A) Relative mRNA expression of ATF2, ATF3, and ATF7 in ABC vs GCB DLBCL biopsies. Error bars indicate standard error of the means (SEMs). (B) Immunofluorescence staining of ATF3 expression and localization in the indicated GCB and ABC DLBCL cell lines. Bar, 5 μm. (C) Histologic staining of ATF3 expression in representative biopsy samples of DLBCL patients. Bar, 50 μM; magnification, ×400. (D-E) TMA analysis of DLBCL biopsies previously categorized into GCB or non-GC using the Hans algorithm (D), or into ABC or GCB by GEP (E). Bar graphs summarizing the classification of non-GC and GCB patients (D) or ABC and GCB patients (E) according to staining intensity (weak, intermediate [int.], and strong) and subcellular localization of ATF3 (C, cytoplasmic; N, partially or predominantly nuclear). Analysis was performed on 28 (D) or 17 (E) patients with nodal (lymph node) tumors. DAPI, 4,6 diamidino-2-phenylindole.
Discussion
In the present study, we demonstrated a new and essential role for the activation of the Jun/ATF branch of the AP-1 pathway in the growth of ABC DLBCL, and identified strong ATF3 expression as a hallmark of this cancer. When exploring the molecular causes of AP-1 activation in ABC DLBCL, we observed that c-Jun, JunB, and JunD levels were systematically upregulated in ABC DLBCL cell lines, and that c-Jun and JunB upregulation occurred in a CARMA1/MALT1- or MyD88/IRAK-1–dependent manner. Recent studies have shown that CARMA1 can drive c-Jun and JunB expression,18 and promote AP-1 activation via the adaptor protein BCL10, which recruits MEKK7 to promote JNK2-mediated c-Jun phosphorylation and stabilization.47 We extend these findings and show that constitutive activation of MyD88 and IRAK also contributes to the increased expression of these AP-1 family members in ABC DLBCL cell lines. However, and consistent with a recent study,39 we saw no correlation of JNK activation with c-Jun upregulation in ABC DLBCL (except for HBL-1 cells). Instead, we found that JunB expression and c-Jun phosphorylation and expression were efficiently blocked by inhibition of the Ser/Thr kinase TAK1. Therefore, TAK1 may directly or indirectly control c-Jun and JunB stability.
Using biochemical approaches, we explored the molecular composition of AP-1 complexes in DLBCL cell lines, and identified ATF2, ATF7, and ATF3 as specific constitutive binding partners of c-Jun, JunB, and JunD. Interestingly, c-Jun/ATF2- and c-Jun/ATF7-containing complexes were generally much more abundant in ABC DLBCL cell lines, and ATF3-containing AP-1 heterodimers with c-Jun, JunB, or JunD were identified as a selective feature of ABC DLBCL cell lines. Using a dominant-negative A-Fos construct or shRNA-mediated silencing of individual AP-1 components, we demonstrated that Jun/ATF-type complexes are important drivers of the proliferation of DLBCL cell lines. These findings are consistent with a previously proposed role for Jun/ATF complexes in conducting autocrine cell growth in other malignancies.48
In the present study, we have attained a better understanding of the molecular mechanisms driving high expression of ATF3 in ABC DLBCL. An important aspect of the regulation of ATF3 levels was its transcriptional upregulation in ABC DLBCL, which was controlled by c-Jun and ATF2. Indeed, the ATF3 promotor has been previously shown to be a c-Jun/ATF2-target in HeLa cells.49 Therefore, high levels of ATF3 are most likely maintained in ABC DLBCL cells by a positive feedback loop. Additionally, our finding of constitutive phosphorylation of JDP2 in ABC as compared with GCB DLBCL cell lines may provide another explanation for the increased levels of ATF3 in the ABC DLBCL subtype. JDP2 binds the ATF3 promotor and suppresses ATF3 transcription, and phosphorylation has been reported to control JDP2 protein stability.42 Understanding of the mechanism underlying constitutive JDP2 phosphorylation and turnover, and of its relevance for cellular transformation in ABC DLBCL will be an interesting aspect of future studies.
ATF3 has been described to have controversial roles in either oncogenesis or tumor suppression in diverse tumor models.50 These seemingly conflicting roles depend on the type of ATF3 homo- or heterodimers formed, which have different DNA-binding specificities, and thus on the expression and activity of individual ATF3 binding partners in the cellular context.50 In ABC DLBCL, ATF3 was highly expressed and constitutively bound to c-Jun, JunB or JunD, suggesting an important role for Jun/ATF3 heterodimers in lymphomagenesis that is consistent with several previous studies highlighting a proliferation-promoting and/or cell-transforming role for ATF3.51-54 We consistently observed a tendency toward strong and nuclear ATF3 expression in samples from patients with ABC DLBCL that were classified either by the Hans algorithm or by GEP. Interestingly, high constitutive ATF3 expression has also been described for Hodgkin lymphoma cells, which depend on ATF3 expression for viability,55 and in adult T-cell leukemia cells,56 suggesting that ATF3 promotes different types of lymphomas by a common mechanism. In conjunction with these data, our findings support an important oncogenic role for ATF3 expression in ABC DLBCL that could be of diagnostic relevance and may inspire novel therapeutic strategies to interfere with ABC DLBCL by targeting the function and/or expression of specific AP-1 family members.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Cristian Smulski for technical advice, Slavica Masina for grammatical revision of the manuscript, Manfredo Quadroni (Protein Analysis Facility, Center for Integrative Genomics, University of Lausanne) for sample analysis by mass spectrometry, and Prof Inti Zlobec and the team of the Translational Research Unit of the Institute of Pathology at the University of Bern for technical assistance with the immunohistochemical staining of human samples.
This work was supported by grants of the Swiss Cancer League and the Stiftung zur Krebsbekämpfung (M.T.) and a Swiss National Science Foundation (SNSF) Sinergia grant (CRSII3_147620 [M.T. and G.L.]). G.L. acknowledges additional support by Deutsche Krebshilfe, the Else-Kröner-Fresenius Stiftung, and the Deutsche Forschungsgemeinschaft (DFG EXC 1003 Cells in Motion).
Authorship
Contribution: M.J. conceived and designed the project, performed experiments, analyzed data, and wrote the paper; M. Gonzalez, Z.J., Y.B., and T.E. performed experiments, analyzed data, and revised the manuscript; U.N. and A.T. provided clinical samples and revised the manuscript; S.H. contributed to the study design and revised the manuscript; G.L. contributed to the study design, provided clinical samples, analyzed data, and revised the manuscript; M. Grau provided statistical analysis of mRNA expression data from a patient cohort; and M.T. conceived and designed the project, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for S.H. is Interfaculty Institute for Biochemistry, University of Tübingen, Tübingen, Germany.
Correspondence: Margot Thome, Department of Biochemistry, University of Lausanne, Chemin des Boveresses 155, CH-1066 Epalinges, Switzerland; e-mail: margot.thomemiazza@unil.ch.

![Figure 5. Strong nuclear ATF3 expression is a hallmark of non-GC and ABC DLBCL patients. (A) Relative mRNA expression of ATF2, ATF3, and ATF7 in ABC vs GCB DLBCL biopsies. Error bars indicate standard error of the means (SEMs). (B) Immunofluorescence staining of ATF3 expression and localization in the indicated GCB and ABC DLBCL cell lines. Bar, 5 μm. (C) Histologic staining of ATF3 expression in representative biopsy samples of DLBCL patients. Bar, 50 μM; magnification, ×400. (D-E) TMA analysis of DLBCL biopsies previously categorized into GCB or non-GC using the Hans algorithm (D), or into ABC or GCB by GEP (E). Bar graphs summarizing the classification of non-GC and GCB patients (D) or ABC and GCB patients (E) according to staining intensity (weak, intermediate [int.], and strong) and subcellular localization of ATF3 (C, cytoplasmic; N, partially or predominantly nuclear). Analysis was performed on 28 (D) or 17 (E) patients with nodal (lymph node) tumors. DAPI, 4,6 diamidino-2-phenylindole.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/127/14/10.1182_blood-2015-07-655647/4/m_1780f5.jpeg?Expires=1767752309&Signature=Z9BMxBhzVrU3NcExpctuUp2DRGQDyBzBVFKa5N4JKJfAEFblRj6jdTdIOpxCg45yZHke~pmD9Kvaa025QzbFJQpKPCPlcG6aT2FBK0NBcjKn8bVai3J7rBuc~ljjdLR8Iuz3CTtlDaukRpMNjArbzcygKHKmOF0EdszOoLoU2tdAg-NAUM7MAhdzQksjINvDHdjtC6DGhILqtXL-rL1N2N0MaOaxbD2Fz7tnZb4Fa5UB4~xt332e9S162DQ72Mlg~-3g9HP6GsuAvBVWUAo2~lU8lh63LQw6VzBz-Wzw~Jaxqeu9QZeqfSwfSb0sRz9-pFBT01W5xZzk7g77SB3YSQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal