Key Points
Glycosylation at N1574 stabilizes the VWF A2 domain against unfolding and proteolysis by ADAMTS13, and its first GlcNAc is the critical element.
Y1544 is a likely interacting residue with N1574-GlcNAc, and its mutation to aspartic acid stabilizes the domain in the absence of the glycan.
Abstract
Shear forces in the blood trigger a conformational transition in the von Willebrand factor (VWF) A2 domain, from its native folded to an unfolded state, in which the cryptic scissile bond (Y1605-M1606) is exposed and can then be proteolysed by ADAMTS13. The conformational transition depends upon a Ca2+ binding site and a vicinal cysteine disulfide bond. Glycosylation at N1574 has previously been suggested to modulate VWF A2 domain interaction with ADAMTS13 through steric hindrance by the bulky carbohydrate structure. We investigated how the N-linked glycans of the VWF A2 domain affect thermostability and regulate both the exposure of the ADAMTS13 binding sites and the scissile bond. We show by differential scanning fluorimetry that the N-linked glycans thermodynamically stabilize the VWF A2 domain. The essential component of the glycan structure is the first sugar residue (GlcNAc) at the N1574 attachment site. From its crystal structures, N1574-GlcNAc is predicted to form stabilizing intradomain interactions with Y1544 and nearby residues. Substitution of the surface-exposed Y1544 to aspartic acid is able to stabilize the domain in the absence of glycosylation and protect against ADAMTS13 proteolysis in both the VWF A2 domain and FLVWF. Glycan stabilization of the VWF A2 domain acts together with the Ca2+ binding site and vicinal cysteine disulfide bond to control unfolding and ADAMTS13 proteolysis.
Introduction
Von Willebrand factor (VWF) is one of the largest circulating proteins of the vasculature. A mature VWF monomer (∼250 kDa) is synthesized by endothelial cells and megakaryocytes with the domain structure D′-D3-A1-A2-A3-D4-C1-C2-C3-C4-C5-C6-CK.1 The large size of VWF observed (upwards of 20 000 kDa) is due to the multimeric/concatemeric nature, arising from intermonomer disulfide linkages between the D′-D3 to D′-D3 and CK to CK domains of neighboring VWF monomers.2 Other important post-translational modifications include the addition of 12 N-linked and 10 O-linked glycans to the mature protein.3 VWF is packaged into the Weibel-Palade bodies of endothelial cells and the α-granules of platelets. VWF is constitutively secreted into the vasculature by endothelial cells, and when stressed, the endothelium and platelets are induced to secrete their VWF-rich vesicles.
The functions of VWF in the vasculature are determined by its conformational state and its multimeric size. Platelet binding to the VWF A1 domain and ADAMTS13 cleavage of the VWF A2 domain depend upon exposure of functional binding sites that are hidden under quiescent conditions.4 These sites are conformationally activated at the molecular level by mechanical force. Crystallization and single-molecule pulling experiments have elucidated the subtle changes that occur in the VWF A1 domain structure that place it in a favorable conformation to interact with the platelet cell-surface receptor GpIBα5-8 and more dramatic changes that result from unraveling of the VWF A2 domain to facilitate ADAMTS13 cleavage.9 In vivo, the activating mechanical forces are applied to VWF through the rheological and shear forces in the vasculature, with increased intramolecular forces applied upon ultralarge VWF multimers.10 Naturally occurring mutations in the VWF gene result in defects in the VWF protein, perturbing these activation mechanisms leading to qualitative defects in VWF function causing the bleeding disorder von Willebrand disease (VWD).11
The multimeric size of VWF in the plasma is regulated by the metalloprotease ADAMTS13,12 which reduces the size of the highly thrombotic ultralarge VWF multimers and prevents spontaneous unfolding and platelet capture. The key regulator of this axis is the binding to and proteolysis of the VWF A2 domain by ADAMTS13. The VWF A2 domain is unique within VWF, as it is the only domain that lacks multiple intradomain or a domain-spanning disulfide bond(s). The structure, function, and docking points of ADAMTS13 with VWF during proteolysis have been well defined at the molecular level.12-14 While it has been demonstrated (by single-molecule techniques) that the VWF A2 can both unfold and refold under mechanical force,9,15,16 the factors regulating the transition from folded to unfolded state have not been fully elicited.
The crystal structure of the VWF A2 domain has demonstrated that the scissile bond (Y1605-M1606) is buried within the core of the domain in its native state and inaccessible to ADAMTS13.15,17,18 Biochemical and biophysical studies have revealed that the vicinal disulfide bond19 (C1669-C1670, VicCC) and Ca2+ binding site (CBS) 15,16,18,20 (D1498, D1596, N1602) of the VWF A2 domain stabilize the domain, provide resilience to unfolding and protect from ADAMTS13 proteolysis. Recently, we demonstrated that both the VicCC and CBS control the susceptibility of full-length VWF (FLVWF) to ADAMTS13 proteolysis.20
N-linked glycosylation of the VWF A2 domain has also been shown to modulate interaction with ADAMTS13,21 but with an unclear mechanism. In this report, we demonstrate that the N1574-linked glycan stabilizes the VWF A2 domain independently of the CBS. The first GlcNAc of the glycan is essential for stabilization.
Methods
Recombinant VWF protein expression and characterization
The recombinant VWF A2 domain, WT VWF A2 (amino acids 1473-1670), with a C-terminal myc-His tag was mutated and expressed, and VWF A2 proteins (wild-type [WT] and variants) were purified by Ni2+ chromatography and quantified by bicinchoninic acid assay, as previously described.20 Mutations were introduced in pcDNA3.1-VWF using KOD polymerase (EMD Millipore), according to the manufacturer’s instructions. To obtain glycan structural variants, the VWF A2 domain was expressed in the HEK293 GnT−/− deficient cell line22 (a kind gift from Dr P. Reeves, University of Essex) and purified as previously described.20 Enzymatic digestion by recombinant PNGase F (New England Biolabs) was performed on WT VWF A2 expressed and purified from HEK293 EBNA cells. EndoH (New England Biolabs) digestion was performed on WT VWF A2 expressed and purified from HEK293 Gnt−/− cells. After digestion, proteins were repurified. Final concentrations were determined using the bicinchoninic acid assay. Proteins were analyzed by 4% to 12% Bis-Tris sodium dodecyl sulfate (SDS) gel followed by silver stain to check purity and molecular weight (MW).
FLVWF and its variants (N1574Q, Y1544D, and Y1544D/N1574Q) were expressed in HEK293 cells and concentrated and dialyzed in 20 mM Tris (pH 7.8) 150 mM NaCl. Multimer gels were performed as previously described.20
Differential scanning fluorimetry
Differential scanning fluorimetry (DSF) assays were carried out as previously described.20,23 In brief, samples contained 2 μg purified VWF A2 domain, 5× SyproOrange dye (Sigma), 1 mM EDTA, or 5mM CaCl2 in 20 mM Tris (pH 7.8), 50 mM NaCl in a total volume of 60 μL. The fluorescence increase brought about by unfolding of the VWF A2 domain was monitored by high‐resolution melting analysis using Rotor‐Gene Q real‐time polymerase chain reaction instrument (Qiagen).
Solution competition binding between ADAMTS13 and VWF A2 variants
Recombinant ADAMTS13 and its inactive variant, E225A (iADAMTS13), with a C-terminal myc/his tag was stably expressed in HEK293 cells as previously described.20 Next, 50 nM purified WT VWF A2 was coated on a 96-well Maxisorp plate (Nunc) overnight at 4°C, and 30 nM iADAMTS13 was separately preincubated with varying concentrations of VWF A2 variants (0-500 nM) in the presence of 5 mM CaCl2 or 1 mM EDTA at 37°C for 1 hour. After blocking (Tris-buffered saline [TBS], 0.1% Tween, 3% bovine serum albumin), the plates were washed with an appropriate buffer (TBS, 0.1% Tween, 5 mM CaCl2 or TBS, 0.1% Tween, 1 mM EDTA) and the preincubated VWF A2/iADAMTS13 added to the plate for 1 to 2 hours. After subsequent wash steps, bound iADAMTS13 was detected using a biotinylated antibody to ADAMTS13, as previously described.24 Appropriate controls were used to determine the solution binding of iADAMTS13 to the VWF A2 domain variants during the preincubation step. The 100% solution binding control contained 0 nM iADAMTS13, whereas the 0% solution binding control contained 0 nM VWF A2. The percent solution binding of VWF A2 titrations was derived from the known optical density values of 0% and 100% controls, and binding curves were fitted using the 1-site binding equation (GraphPad Prism).
ADAMTS13 proteolysis of VWF A2 and FLVWF
The VWF A2 domain variants (2 µM), FLVWF variants (1 µg/mL), and ADAMTS13 were separately preincubated in 20 mM Tris (pH 7.8) 150 mM NaCl, 5 mM CaCl2 ± 2 M urea at 37°C for 45 minutes. ADAMTS13 was added to the VWF variants at a final concentration of 20 nM and incubated at 37°C for proteolysis to occur. Reactions were terminated by addition of EDTA to a final concentration of 100 mM. To determine proteolysis of the VWF A2 domain variants, SDS loading buffer (Invitrogen) was added, and samples were heated at 95°C for 10 minutes and then loaded onto a 4% to 12% Bis-Tris gel followed by silver staining. To determine proteolysis of FLVWF, reducing regent (Invitrogen) and SDS loading buffer (Invitrogen) were added and samples were heated at 70°C for 30 minutes. Reduced samples were run on a 3% to 8% Tris-acetate gel and intact VWF and its proteolytic fragment detected by western blot using a cocktail of VWF polyclonal (Dako) and VWF-ADAMTS13 cleaved monoclonal antibodies (R&D Systems; #2764-WF).
Results
The N1574-GlcNAc linkage stabilizes the VWF A2 domain
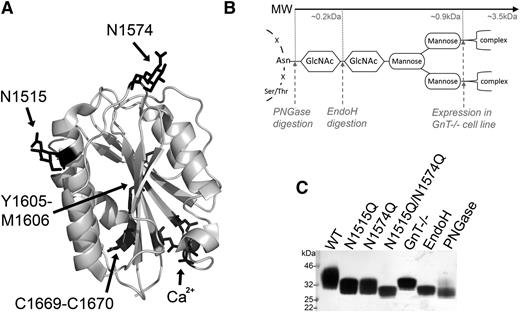
The crystal structure of the VWF A2 domain shown in Figure 1A highlights the structural features of the domain: the scissile bond buried at the core (Y1605-M1606), the C-terminal vicinal disulfide bond (C1669-C1670), the calcium binding site (Ca2+: D1498, D1596, N1602), and the 2 N-linked glycans (N1515 and N1574). To assess the functional role of glycosylation of the VWF A2 domain, we expressed the VWF A2 domain with both glycans attached (WT) or prevented N-linked glycosylation through mutation of the asparagine attachment sites (N1515Q, N1574Q, and N1515Q/N1574Q). To further investigate the function of the potentially large and bulky carbohydrate glycan structure (Figure 1B), selective enzymatic trimming (with EndoH and PNGase, the latter resulting in conversion of Asn to Asp) and inhibition of complex glycans to the trimannose core (by expression in HEK GnT−/− cells) were used. Successful removal/trimming of the VWF A2 domain variants is clearly demonstrated by the reduction of MW of the variants (Figure 1C).
Structure and manipulation of the VWF A2 domain N-linked glycans. (A) Crystal structure of the VWF A2 domain (3ZQK, chain C) is shown with the 2 N-linked glycosylation attachment sites (N1515 and N1574), scissile bond (Y1605-M1606), vicinal disulfide bond (C1669-C1670) and calcium binding site (Ca2+) residues in black. (B) Basic representation of N-linked glycan structure with asparagine backbone attachment motif and carbohydrate structure shown in black. Enzymatic cleavage sites (PNGase and EndoH) and inhibition of complex sugars (GnT−/−) shown by gray dashed lines with approximate MWs displayed. (C) VWF A2 domain fragments with full glycosylation (WT), mutated asparagine attachment site(s) (N1515, N1574Q, and N1515/N1574Q), or truncated glycan structures (GnT−/−, EndoH, and PNGase) after expression with or without enzymatic treatment and purification were analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) on a 4% to 12% Bis-Tris gel followed by silver staining to determine changes in MW/glycan structure.
Structure and manipulation of the VWF A2 domain N-linked glycans. (A) Crystal structure of the VWF A2 domain (3ZQK, chain C) is shown with the 2 N-linked glycosylation attachment sites (N1515 and N1574), scissile bond (Y1605-M1606), vicinal disulfide bond (C1669-C1670) and calcium binding site (Ca2+) residues in black. (B) Basic representation of N-linked glycan structure with asparagine backbone attachment motif and carbohydrate structure shown in black. Enzymatic cleavage sites (PNGase and EndoH) and inhibition of complex sugars (GnT−/−) shown by gray dashed lines with approximate MWs displayed. (C) VWF A2 domain fragments with full glycosylation (WT), mutated asparagine attachment site(s) (N1515, N1574Q, and N1515/N1574Q), or truncated glycan structures (GnT−/−, EndoH, and PNGase) after expression with or without enzymatic treatment and purification were analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) on a 4% to 12% Bis-Tris gel followed by silver staining to determine changes in MW/glycan structure.
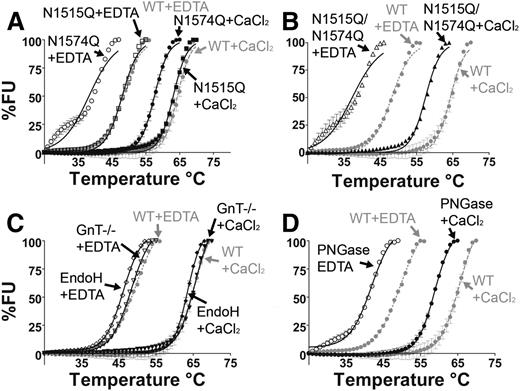
The purified VWF A2 glycan variants were examined by DSF assay to evaluate stability to thermal unfolding. The normalized temperature induced unfolding curves of the VWF A2 glycan variants in both the presence of Ca2+ (5 mM CaCl2) and absence of Ca2+ (1 mM EDTA) are displayed in Figure 2. It has previously been determined that Ca2+ stabilizes the VWF A2 domain in a concentration-dependent manner, with a dissociation constant (KD) of 0.2µM15 and 3.8µM.16 In the presence of 1mM EDTA, WT VWF A2 shows a reduction in thermostability in comparison with WT+ CaCl2 (Figure 2A-D, gray curves) with a transition midpoint (Tm) of 48.2°C ± 0.05°C and 64.3°C ± 0.13°C respectively, consistent with previous results.15,20
N-linked glycosylation stabilizes the VWF A2 domain through the N1574-GlcNAc linkage. DSF measurements, expressed as percent fluorescence (% FU), were carried out in the presence of 1 mM EDTA (open symbols) or 5 mM CaCl2 (closed symbols). WT VWF A2 was studied (gray lines, A-D) as well as VWF A2 glycan variants N1515Q, N1574Q (A), and N1515Q/N1574Q (B) and variants prepared in HEK293 GnT−/− deficient cells and following digestion with EndoH (C) and PNGase (D). Results are means ± standard error of the mean (SEM) of at least 3 independent experiments.
N-linked glycosylation stabilizes the VWF A2 domain through the N1574-GlcNAc linkage. DSF measurements, expressed as percent fluorescence (% FU), were carried out in the presence of 1 mM EDTA (open symbols) or 5 mM CaCl2 (closed symbols). WT VWF A2 was studied (gray lines, A-D) as well as VWF A2 glycan variants N1515Q, N1574Q (A), and N1515Q/N1574Q (B) and variants prepared in HEK293 GnT−/− deficient cells and following digestion with EndoH (C) and PNGase (D). Results are means ± standard error of the mean (SEM) of at least 3 independent experiments.
Mutation of the most N-terminal glycosylation site (N1515Q) caused no apparent effect on the thermostability of the VWF A2 domain (Figure 2A and Table 1). In contrast, mutant N1574Q (Figure 2A) and double mutation N1515Q/N1574Q (Figure 2B) caused a reduction in Tm of 10°C to 11°C in 1 mM EDTA and 6.8°C in 5 mM CaCl2 (Table 1), indicating a reduction in thermostability. Glycosylation at N1574 (but not N1515) therefore imparts an independent stabilizing effect on the VWF A2 domain in a manner distinct from that of Ca2+.
Tm of VWF A2 domain variants as determined by DSF
| . | Tm (°C) . | |
|---|---|---|
| 1 mM EDTA . | 5 mM CaCl2 . | |
| WT | 48.2 ± 0.05 | 64.3 ± 0.13 |
| N1515Q | 48.0 ± 0.04 | 63.1 ± 0.07 |
| N1574Q | 38.2 ± 0.17 | 57.5 ± 0.07 |
| N1515Q/N1574Q | 37.3 ± 0.23 | 57.5 ± 0.05 |
| GnT−/− | 47.5 ± 0.04 | 64.3 ± 0.07 |
| EndoH | 45.5 ± 0.04 | 63.0 ± 0.08 |
| PNGase | 40.4 ± 0.11 | 58.3 ± 0.07 |
| R1575V | 47.6 ± 0.06 | 60.9 ± 0.05 |
| R1575V/N1574Q | 39.6 ± 0.08 | 55.4 ± 0.04 |
| Y1544A | 47.9 ± 0.05 | 63.6 ± 0.15 |
| Y1544A/N1574Q | 43.1 ± 0.07 | 60.8 ± 0.16 |
| Y1544D | 53.5 ± 0.07 | 68.5 ± 0.12 |
| Y1544D/N1574Q | 52.4 ± 0.07 | 67.2 ± 0.10 |
| Y1544R | 45.3 ± 0.16 | 63.1 ± 0.10 |
| N1574Q/Y1544R | 32.3 ± 0.52 | 58.8 ± 0.09 |
| N1515Q/N1574Q/Y1544D | 52.7 ± 0.08 | 67.6 ± 0.32 |
| . | Tm (°C) . | |
|---|---|---|
| 1 mM EDTA . | 5 mM CaCl2 . | |
| WT | 48.2 ± 0.05 | 64.3 ± 0.13 |
| N1515Q | 48.0 ± 0.04 | 63.1 ± 0.07 |
| N1574Q | 38.2 ± 0.17 | 57.5 ± 0.07 |
| N1515Q/N1574Q | 37.3 ± 0.23 | 57.5 ± 0.05 |
| GnT−/− | 47.5 ± 0.04 | 64.3 ± 0.07 |
| EndoH | 45.5 ± 0.04 | 63.0 ± 0.08 |
| PNGase | 40.4 ± 0.11 | 58.3 ± 0.07 |
| R1575V | 47.6 ± 0.06 | 60.9 ± 0.05 |
| R1575V/N1574Q | 39.6 ± 0.08 | 55.4 ± 0.04 |
| Y1544A | 47.9 ± 0.05 | 63.6 ± 0.15 |
| Y1544A/N1574Q | 43.1 ± 0.07 | 60.8 ± 0.16 |
| Y1544D | 53.5 ± 0.07 | 68.5 ± 0.12 |
| Y1544D/N1574Q | 52.4 ± 0.07 | 67.2 ± 0.10 |
| Y1544R | 45.3 ± 0.16 | 63.1 ± 0.10 |
| N1574Q/Y1544R | 32.3 ± 0.52 | 58.8 ± 0.09 |
| N1515Q/N1574Q/Y1544D | 52.7 ± 0.08 | 67.6 ± 0.32 |
Results are means ± SEM of at least 3 independent experiments.
The truncated glycosylation variants (termed GnT−/−, EndoH, and PNGase after their means of preparation) were subjected to DSF assays (Figure 2 C-D). Truncation of the glycan structure to the trimannose core (GnT−/−) and selective trimming to the first GlcNAc sugar (EndoH) caused minimal divergence from the unfolding curves of fully glycosylated WT VWF A2 (Figure 2C). Comparison of the Tm of the partial glycan structural variants (GnT−/− and EndoH) to that of WT indicated minimal reduction in stability in both 1 mM EDTA and 5 mM CaCl2 (Table 1). In contrast, complete enzymatic removal of glycosylation (termed PNGase) resulted in a reduction in thermostability in both 1 mM EDTA and 5 mM CaCl2 (Figure 2D), causing a shift in Tm from WT of 7.8°C in 1 mM EDTA and 6°C in 5 mM CaCl2 (Table 1). It is therefore evident that the complex glycans of the VWF A2 domain are not essential for domain stabilization. The key component of N-linked glycosylation for stability of the VWF A2 domain appears to be the initial N1574-GlcNAc residue, which is retained on the EndoH-treated variant.
Removal of glycan at N1574 increases solution binding of the VWF A2 to ADAMTS13
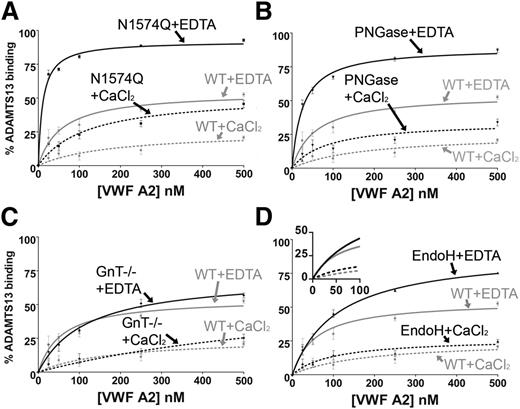
WT VWF A2 was immobilized on the surface of a microtiter plate, causing it to unfold and present its ADAMTS13 binding sites. In previous reports, WT ADAMTS13 was preincubated with VWF fragments in the presence of 10 mM EDTA to prevent proteolysis and recycling of the protease.24 Although EDTA inhibits ADAMTS13 proteolysis of VWF, it also prevents the Ca2+-dependent stabilization of the domain. In the present study, to prevent proteolysis in the presence of Ca2+, inactive ADAMTS13 (iADAMTS13,25 with the E225A mutation) was used to bind to immobilized WT VWF A2 and the in-solution competitive glycan variants.
Preincubation of the VWF A2 glycan variants with 30 nM iADAMTS13 was carried out in both 1 mM EDTA and 5 mM CaCl2. The WT VWF A2 domain displayed a ∼2.5-fold enhancement of solution binding in the competition assay in the presence of 1 mM EDTA at saturating concentrations (500 nM) of the VWF A2 domain in comparison with the WT VWF A2 domain in the presence of 5 mM CaCl2 (Figure 3A-D). The presence of Ca2+ therefore stabilizes the VWF A2 domain and protects against solution binding of ADAMTS13.
Solution competition assay of ADAMTS13 with VWF A2 domain glycan variants. WT VWF A2 domain (50 nM) was immobilized on a microtiter plate. A total of 30 nM (inactive) iADAMTS13 (E225A) was separately preincubated with varying concentration of the VWF A2 domain glycan variants (0-500 nM) in the presence of 1 mM EDTA or 5mM CaCl2 for 1 hour at 37°C: N1574Q (A), PNGase (B), GnT−/− (C), and EndoH (D). The preincubated VWF A2/ADAMTS13 was added to the microtiter plate for 1 or 2 hours, where the preincubated VWF A2 domain variants compete with the immobilized WT VWF A2 domain for binding to the iADAMTS13. After washing, iADAMTS13 bound to the immobilized VWF A2 domain was detected by a biotinylated anti–TSP2-4 polyclonal antibody followed by streptavidin–horseradish peroxidase and o-phenylenediamine hydrochloride. Preincubation controls containing 0 nM VWFA2 and 30 nM iADAMTS13 represented 0% solution binding of the VWF A2 domain to iADAMTS13, and 0 nM VWF A2 and 0 nM iADAMTS13 represented 100% solution binding of VWF A2 to ADAMTS13. Results were plotted as percent solution binding against preincubation concentration of the VWF A2 domain variant and fitted using the 1-site binding model (GraphPad). Results are means ± SEM of at least 3 independent experiments.
Solution competition assay of ADAMTS13 with VWF A2 domain glycan variants. WT VWF A2 domain (50 nM) was immobilized on a microtiter plate. A total of 30 nM (inactive) iADAMTS13 (E225A) was separately preincubated with varying concentration of the VWF A2 domain glycan variants (0-500 nM) in the presence of 1 mM EDTA or 5mM CaCl2 for 1 hour at 37°C: N1574Q (A), PNGase (B), GnT−/− (C), and EndoH (D). The preincubated VWF A2/ADAMTS13 was added to the microtiter plate for 1 or 2 hours, where the preincubated VWF A2 domain variants compete with the immobilized WT VWF A2 domain for binding to the iADAMTS13. After washing, iADAMTS13 bound to the immobilized VWF A2 domain was detected by a biotinylated anti–TSP2-4 polyclonal antibody followed by streptavidin–horseradish peroxidase and o-phenylenediamine hydrochloride. Preincubation controls containing 0 nM VWFA2 and 30 nM iADAMTS13 represented 0% solution binding of the VWF A2 domain to iADAMTS13, and 0 nM VWF A2 and 0 nM iADAMTS13 represented 100% solution binding of VWF A2 to ADAMTS13. Results were plotted as percent solution binding against preincubation concentration of the VWF A2 domain variant and fitted using the 1-site binding model (GraphPad). Results are means ± SEM of at least 3 independent experiments.
To assess the role of N-linked glycosylation of the VWF A2 domain on the solution interaction with ADAMTS13, WT VWF A2 and its domain variants N1574Q, GnT−/−, EndoH, and PNGase were preincubated with 30nM iADAMTS13 at varying concentrations (0-500nM). Mutation of the stabilizing glycan attachment residue (N1574Q) and complete removal of glycosylation by PNGase (N1515D/N1574D) resulted in enhancements of solution binding compared with WT in both 1 mM EDTA and 5 mM CaCl2 (Figure 3A-B). Truncation of N-linked glycosylation to the trimannose core (with the GnT−/− variant) had minimal effect on the solution interaction of the VWF A2 domain with ADAMTS13 (Figure 3C). Although enzymatic trimming to the first GlcNAc sugar (the EndoH variant) shows a similar effect on the solution binding in the presence of 5 mM CaCl2 to that of WT, some enhancement of binding was observed in 1 mM EDTA (Figure 3D). Nevertheless, at concentrations lower than saturating (ie, between 0 and 100 nM), VWF A2 EndoH displayed binding curves similar to that of WT VWF A2 (Figure 3D, inset). Maximal destabilization of the VWF A2 domain implies full unfolding and stoichiometric binding to iADAMTS13, something that is not independently determined here.
To confirm that truncation/removal of N-linked glycosylation from the VWF A2 domain did not disrupt native ADAMTS13 binding sites, the VWF A2 glycan variants were immobilized on a microtiter plate and iADAMTS13 binding was studied. Similar KD(app) values were obtained with WT and variant VWF A2 preparations (data not shown). These results indicate that N-linked glycosylation prevents ADAMTS13 interaction not through steric hindrance but rather through a stabilizing interaction mediated largely through the N1574-GlcNAc sugar.
Intradomain stabilizing interactions of N1574-GlcNAc and Y1544
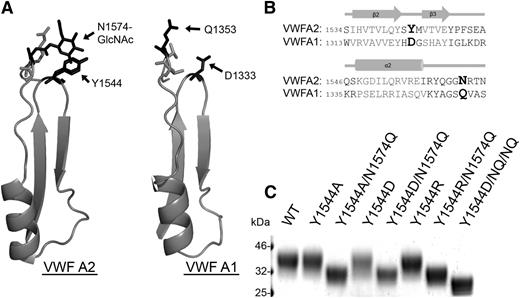
There are 3 tandem von Willebrand A-type domains in the VWF sequence,26 and the VWF A2 domain is the only one that is N-glycosylated. Based on the thermostability (Figure 2) and iADAMTS13 binding results (Figure 3), it can be concluded that the crucial component of VWF A2 domain glycosylation for modulating the interaction with ADAMTS13 is the N1574-GlcNAc linkage rather than the bulky carbohydrate structure. N1574 is located in the α2-α3 loop region of the VWF A2 domain fold, with the N1574 R-group and attached GlcNAc chain surface exposed and oriented away from the center of the domain. Potential stabilizing interactions of N1574-GlcNAc with Y1544 and adjacent residues can be observed in all chains in both available mammalian VWF A2 crystal structures (3GXB, 3ZQK). Y1544 is positioned at the apex of the tight β2-β3 hairpin turn in the VWA fold and ∼4Å away from N1574-GlcNAc in one of the VWF A2 domain structures (Figure 4A). Alignment of the VWF A2 domain with the homologous unglycosylated VWF A1 domain17 (Figure 4A-B) reveals replacement of Y1544 of the A2 domain with the negatively charged aspartic acid in the A1 domain (Figure 4B, bold). The similarly charged, but unglycosylated, glutamine of the VWF A1 domain is in the homologous position of asparagine glycan attachment site in the VWF A2 domain (Figure 4B, bold).
N1574-GlcNAc intradomain interactions. (A) Part of the crystal structures of the regions of the VWF A2 domain (3ZQK, I1535-N1577) and VWF A1 domain (1AUQ, V1314-S1356) containing the β2-β3 hairpin and α2-α3 loop. (B) Alignment of the VWF A2 and VWF A1 domain (adapted from Zhang et al17 ) regions containing important glycosylation site and potential hydrophobic interaction sites. Amino acids forming secondary structures in gray and key residues displayed in larger bold font. (C) VWF A2 domain fragments containing mutation(s) to glycosylation site, a candidate hydrophobic interaction partner, or combinations of these were analyzed by SDS-PAGE on a 4% to 12% Bis-Tris gel followed by silver staining after expression and purification to determine changes in MW caused by alteration of the surface glycan structure (the N1515Q/N1574Q/Y1544D variant is labeled as Y1544D/NQ/NQ).
N1574-GlcNAc intradomain interactions. (A) Part of the crystal structures of the regions of the VWF A2 domain (3ZQK, I1535-N1577) and VWF A1 domain (1AUQ, V1314-S1356) containing the β2-β3 hairpin and α2-α3 loop. (B) Alignment of the VWF A2 and VWF A1 domain (adapted from Zhang et al17 ) regions containing important glycosylation site and potential hydrophobic interaction sites. Amino acids forming secondary structures in gray and key residues displayed in larger bold font. (C) VWF A2 domain fragments containing mutation(s) to glycosylation site, a candidate hydrophobic interaction partner, or combinations of these were analyzed by SDS-PAGE on a 4% to 12% Bis-Tris gel followed by silver staining after expression and purification to determine changes in MW caused by alteration of the surface glycan structure (the N1515Q/N1574Q/Y1544D variant is labeled as Y1544D/NQ/NQ).
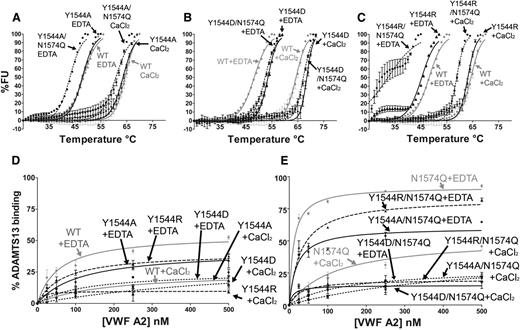
To investigate the potential for stabilizing interactions between Y1544 and N1574-GlcNAc, the following VWF A2 variants were prepared and studied: Y1544A, Y1544A/N1574Q, Y1544D, Y1544D/N1574Q, Y1544R, and Y1544R/N1574Q. The Y1544 and N1574 variants were subjected to DSF assays (Figure 5A-C). Mutation of Y1544, firstly to the smaller hydrophobic Y1544A, resulted in no loss of domain stability in comparison with the WT (Figure 5A and Table 1). Interestingly, combination mutation of Y1544A/N1574Q resulted in an enhancement of thermostability compared with N1574Q alone (Table 1), suggesting that the aromatic tyrosine maybe a source of destabilization. Substitution of Y1544 to aspartic acid rather than alanine in the Y1544D variants resulted in a slight enhancement in thermostability compared with WT VWF A2 domain in both 1 mM EDTA and 5 mM CaCl2 (4.2°C and 2.9°C, respectively; Figure 5B and Table 1). When the Y1544D/N1574Q variant is compared with that of N1574Q, a much larger stabilizing effect is observed, with a 15.3°C increase in 1 mM EDTA and a 11.0°C increase in 5 mM CaCl2; the same stabilizing effect was observed with the N1515Q/N1574Q/Y1544D variant (Figure 4C, Y1544D/NQ/NQ, and Table 1, unfolding curves not shown). Finally, the substitution of Y1544 to the positively charged arginine (Y1544R) resulted in minimal reduction in thermostability in comparison with WT (Figure 5C and Table 1) and is similar in this respect to the Y1544A variant. However, a large decrease in stability was observed with the Y1544R/N1574Q variant (Figure 5C and Table 1), contrasting with the increase in stability when the Y1544 was mutated to the negatively charged aspartic acid (Y1544D/N1574Q).
Functional role of Y1544 in A2 domain stability and ADAMTS13 interaction. VWF A2 variants Y1544A, Y1544A/N1574Q (A), Y1544D, Y1544D/N1574Q (B), and Y1544R and Y1544R/N1574Q (C) were subjected to DSF assay in the presence of 1 mM EDTA or 5 mM CaCl2. Unfolding curves were compared with WT VWF A2 alone (A-C, gray lines). (D-E) The solution competition binding of the above VWF A2 domain variants (Y1544A or Y1544A/N1574Q, circles; Y1544D or Y1544D/N1574Q, squares; Y1544R or Y1544R/N1574Q, triangles) were compared with WT (D, gray lines) and N1574Q (E, gray lines). Results are means ± SEM of at least 3 independent experiments.
Functional role of Y1544 in A2 domain stability and ADAMTS13 interaction. VWF A2 variants Y1544A, Y1544A/N1574Q (A), Y1544D, Y1544D/N1574Q (B), and Y1544R and Y1544R/N1574Q (C) were subjected to DSF assay in the presence of 1 mM EDTA or 5 mM CaCl2. Unfolding curves were compared with WT VWF A2 alone (A-C, gray lines). (D-E) The solution competition binding of the above VWF A2 domain variants (Y1544A or Y1544A/N1574Q, circles; Y1544D or Y1544D/N1574Q, squares; Y1544R or Y1544R/N1574Q, triangles) were compared with WT (D, gray lines) and N1574Q (E, gray lines). Results are means ± SEM of at least 3 independent experiments.
The solution competition binding of these VWF A2 variants with iADAMTS13 was compared with those of WT and N1574Q VWF A2 domains (Figure 5D-E). No enhancement of binding was observed when the Y1544 was mutated either to the uncharged (Y1544A), positively charged (Y1544R) or negatively charged (Y1544D) residues in both 1 mM EDTA and 5 mM CaCl2 (Figure 5D). Y1544 was then mutated in combination with N1574 (Figure 5E). The interaction of Y1544A/N1574Q variant with iADAMTS13 was similar to WT VWF A2 in both EDTA and CaCl2. The Y1544R/N1574Q variant displayed an enhanced solution binding interaction, whereas the Y1544D/N1574Q variant a reduced interaction. This is interpreted as due to an induced repulsive (Y1544R/N1574Q) or stabilizing (Y1544D/N1574Q) interaction with N1574Q. Interactions of the residue at 1544 with the glycan’s neighboring R1575 residue also cannot be excluded.
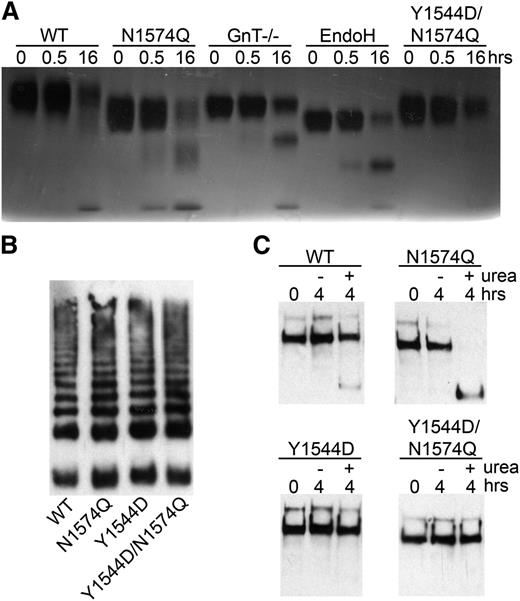
To investigate whether the Y1544D and Y1544D/N1574Q mutations impeded VWF A2 domain unfolding or disrupted native ADAMTS13 binding sites, the mutated VWF A2 domain fragments were coated onto a Maxisorp plate. No difference in KD(app) was observed when comparing to the ADAMTS13 binding of the WT and N1574Q VWF A2 fragments (data not shown). Further to this, the N1574Q/Y1544D mutant was proteolysed by ADAMTS13 (in the presence of 2 M urea) in a manner similar to WT and the glycan variants, although at a slower rate than the latter (Figure 6A).
Proteolysis of glycan variants of the VWF A2 domain and FLVWF. (A) The VWF A2 variants (2 µM) WT, N1574Q, GnT−/−, EndoH, and Y1544D/N1574Q and ADAMTS13 (20 nM) were separately preincubated with 20 mM Tris (pH7.9), 150 mM NaCl, 5 mM CaCl2 and 2 M urea for at 37°C 45 minutes. Samples were combined and incubated at 37°C for proteolysis to occur, and reactions were stopped after 0.5 and 16 hours by the addition of EDTA. Proteolysis was assessed by SDS-PAGE on a 4% to 12% Bis-Tris gel followed by silver stain. (B) FLVWF containing mutation(s) to the stabilizing glycan (N1574Q) or hydrophobic substitution (Y1544D) or combination of (Y1544D/N1574Q) were expressed in HEK 293 cells. Multimer formation was analyzed on a 2% agarose gel and VWF bands detected on a western blot with a polyclonal anti-VWF antibody to determine multimer structure. (C) Proteolysis of FLVWF and its variants by ADAMTS13. FLVWF and its variants (1 µg/mL) and ADAMTS13 (20 nM) were separately preincubated with 20 mM Tris (pH 7.9), 150 mM NaCl, 5 mM CaCl2 ± 2 M urea for at 37°C 45 minutes. Samples were combined and incubated at 37°C for proteolysis to occur, and reactions were stopped after 4 hours by the addition of EDTA. VWF cleavage products were resolved after the addition of reducing reagent. The samples were run on a 3% to 8% Tris-acetate gel and VWF and cleaved VWF bands detected by western blot using a cocktail of anti-VWF polyclonal and monoclonal antibody that detects the C-terminal cleavage product.
Proteolysis of glycan variants of the VWF A2 domain and FLVWF. (A) The VWF A2 variants (2 µM) WT, N1574Q, GnT−/−, EndoH, and Y1544D/N1574Q and ADAMTS13 (20 nM) were separately preincubated with 20 mM Tris (pH7.9), 150 mM NaCl, 5 mM CaCl2 and 2 M urea for at 37°C 45 minutes. Samples were combined and incubated at 37°C for proteolysis to occur, and reactions were stopped after 0.5 and 16 hours by the addition of EDTA. Proteolysis was assessed by SDS-PAGE on a 4% to 12% Bis-Tris gel followed by silver stain. (B) FLVWF containing mutation(s) to the stabilizing glycan (N1574Q) or hydrophobic substitution (Y1544D) or combination of (Y1544D/N1574Q) were expressed in HEK 293 cells. Multimer formation was analyzed on a 2% agarose gel and VWF bands detected on a western blot with a polyclonal anti-VWF antibody to determine multimer structure. (C) Proteolysis of FLVWF and its variants by ADAMTS13. FLVWF and its variants (1 µg/mL) and ADAMTS13 (20 nM) were separately preincubated with 20 mM Tris (pH 7.9), 150 mM NaCl, 5 mM CaCl2 ± 2 M urea for at 37°C 45 minutes. Samples were combined and incubated at 37°C for proteolysis to occur, and reactions were stopped after 4 hours by the addition of EDTA. VWF cleavage products were resolved after the addition of reducing reagent. The samples were run on a 3% to 8% Tris-acetate gel and VWF and cleaved VWF bands detected by western blot using a cocktail of anti-VWF polyclonal and monoclonal antibody that detects the C-terminal cleavage product.
It is concluded that the residue 1544 can influence the stability of the VWF A2 domain. It is positioned to interact (depending on the residue Tyr, Asp or Arg) with residue 1574, which is normally occupied by the glycan.
To study the broader consequences of the N1574-GlcNAc interaction with Y1544, mutations Y1544D, N1574Q, and Y1544D/N1574Q were introduced into FLVWF. The FLVWF variants were transiently expressed in HEK293 cells. All variants were expressed at similar levels (data not shown) and displayed normal multimer structure (Figure 6B). The FLVWF variants were incubated with 20 nM ADAMTS13 in the presence or absence of 2 M urea. Proteolysis assays were terminated after 4 hours, samples reduced, and product detected on a western blot. Under these conditions, partial proteolysis was observed for WT only in the presence of 2 M urea, as indicated by the appearance of a faint band of a lower MW (Figure 6C). Mutation of N1574Q enhanced the proteolysis of FLVWF in the presence of urea, as expected. No proteolysis was observed with the Y1544D and Y1544D/N1574Q mutations, supporting the results of reduced ADAMTS13 interaction and increased stability observed for these variants in the isolated VWF A2 domain.
Discussion
The N-linked glycans of the VWF A2 domain, particularly N1574, have previously been shown to modulate the interaction with ADAMTS13, as demonstrated through proteolysis and binding assays.21 Here, we present a mechanistic study on the glycans of the VWF A2 domain, focusing on their role in its structure and unfolding. Utilizing thermal unfolding and solution binding assays, we have found that the N1574 glycosylation site stabilizes the domain against unfolding and ADAMTS13 binding. Mutation N1574Q causes a 6.8°C reduction in thermostability in comparison with the WT VWF A2 domain in 5 mM CaCl2. Interestingly, the destabilization resulting from mutation of this glycosylation site is quantitatively similar to that observed by mutation of the vicinal disulfide bond and disruption of the calcium binding site of the VWF A2 domain.20 This triad (N-linked glycosylation at N1574, vicinal disulfide bond C1669-C1670, and CBS) of structural features of the VWF A2 domain have all now been demonstrated to stabilize thermodynamically the VWF A2 domain while also modulating its interaction with ADAMTS13.
To further define the mechanism by which N-linked glycosylation of the VWF A2 domain both stabilizes the domain and modulates interaction with ADAMTS13, truncation variants of the glycan structure were made. One of the dominant features of N-linked glycosylation is the large branched carbohydrate structures they form, which can be 3 to 4 kDa per glycosylation site. In the present study, trimming down of the VWF A2 domain glycan structure was performed through expression in GnT−/− cells and by enzymatic digestion with EndoH to the first sugar residue (GlcNAc). The resultant glycan structures show a large decrease in overall size and MW (predicted to be 75% to 95% reduction of the glycan) while still maintaining covalent attachment to the protein backbone. These truncated VWF A2 domain variants displayed characteristics similar to the fully glycosylated WT VWF A2 domain in thermal unfolding and ADAMTS13 binding/cleavage, indicating that the complex glycans that form the majority of the glycan structure27 are not required for modulating the VWF A2/ADAMTS13 interaction.
To gain insight into the importance of the N1574-GlcNAc linkage, it was visualized within the available 3-dimensional structures of the WT VWF A2 domain. Three crystal structures of the VWF A2 domain have been published to date, and 2 of these have been produced in mammalian cells15,17 (using GnT−/− cells and/or EndoH digestion), allowing for visualization of the N1574-GlcNAc linkage. The other structure was produced in bacterial cells18 and was therefore not glycosylated; stabilization was achieved by an engineered disulfide bond connecting the N and C termini and the VWF A2 domain. Utilizing the 2 crystal structures of the mammalian VWF A2 domain, the position and orientation of the N1574-GlcNAc was inspected. The roles of glycans in crystal structures is often obscured by the truncated nature of the glycan and the “snapshot” of the flexible carbohydrate side chain.28 Nevertheless, evidence of stabilizing interactions with the surface-exposed Y1544 was strongly suggested. Y1544 is positioned in the apex of the class 1:3 β-hairpin turn29 observed in the VWA fold. It can be seen in the 3ZQK and 3GXB structures to form hydrogen bonding interactions with the O4 atom of N1574-GlcNAc. In contrast, the unglycosylated bacterial derived structure (3PPV) lacks this feature and as a possible consequence displays flexibility by rotation around the Gly1572-Gly1573 bond.
To obtain additional evidence of intradomain interactions, the candidate interacting Y1544 residue was mutated. β-Hairpins are suggested to be the nucleation sites for protein folding.30 Structural alignment of the VWF A2 domain (3ZQK, chain C) to other known VWA-type folds (http://www.ebi.ac.uk/thornton-srv/databases/sas/) indicates that the large aromatic Y1544 is apparently anomalous in the tight turn region. Tyrosine at position 1544 may be another structural specialization of the VWF A2 domain to allow the potentially reversible transition from folded to unfolded state. We speculate that the correct β-hairpin interaction with α2-β3 loop is only formed in the presence of the N1574-GlcNAc. In the absence of the stabilizing interaction from the N1574-glycan, Y1544 causes local destabilization of the β-hairpin turn, which in turn leads to destabilization of the VWF A2 domain as a whole. Substitution to the smaller, more inert Y1544A maintains a stable folded β-hairpin in the absence of glycosylation at N1574. The enhancement of stability seen in the Y1544D/N1574Q variant (Table 1 and Figure 5B) most likely occurs through a stabilizing interaction with N1574Q or its neighboring residues, which also protects from ADAMTS13 binding/proteolysis (Figures 5D and 6). This suggestion is supported by the decrease in stability seen with the positively charged Y1544R/N1574Q variant when compared with Y1544D/N1574Q (Table 1 and Figure 5B-C).
It is known that mechanical force is required to unfold the VWF A2 domain. This has been determined by AFM to be ∼10 pN.9 Exactly how this force causes a transition in the folded VWF A2 domain to an unfolded state and whether or not an intermediary step exists have not yet been fully elucidated at the molecular level. Thermal15,19,20 and chemical31 unfolding of the VWF A2 domain is a useful tool to probe the structural integrity of the VWF A2 domain. Our results in conjunction with published biophysical studies indicate that there is a clear relationship between the stability of the VWF A2 domain and its propensity to interact with and be cleaved by ADAMTS13.
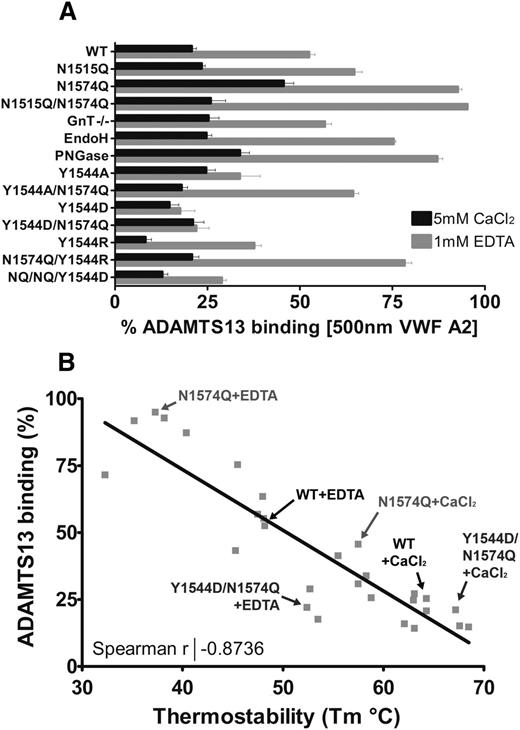
The consequences of modification or removal of the glycans from the VWF A2 domain in terms of its solution binding of ADAMTS13 can be seen in Figure 7A. Of note is the large stabilizing influence of residue Y1544 when mutated to aspartic acid, in the absence of the glycan at N1574. Plotting the condition-dependent (1 mM EDTA or 5 mM CaCl2) Tm of the VWF A2 domain variants against the condition/variant matched solution binding to ADAMTS13 at 500 nM reveals a strong negative correlation (Figure 7B). The more stable the VWF A2 domain is, the less likely it is to interact with ADAMTS13. The data points represented on Figure 7B are of the engineered mutations to the VWF A2 domain. It has previously been demonstrated that VWD type 2A mutations in the isolated VWF A2 domain also cause reduction in thermostability.32 It is likely that the structural defects caused by the naturally occurring VWD type 2A group II mutants reduce the stability of the domain and lower the force threshold at which the VWF A2 domain unfolds, resulting in increased ADAMTS13 proteolysis and reduction in the high-MW VWF multimer species under conditions in which the WT would be resistant to ADAMTS13 proteolysis.
Relationship between VWF A2 domain thermostability and ADAMTS13 binding. (A) The competition binding percentage of the VWF A2 domain variants at saturating concentrations (500 nM) to ADAMTS13 is displayed. Results are means ± SEM of at least 3 independent experiments (the N1515Q/N1574Q/Y1544D variant is labeled as Y1544D/NQ/NQ). (B) The calculated mean Tm was plotted against the mean solution binding of 500 nM VWF A2 to 30nM iADAMTS13 for the variant and condition matched VWF A2 domain variants. Example data points are indicated on the graph, and the Spearman coefficient was calculated using linear regression (GraphPad).
Relationship between VWF A2 domain thermostability and ADAMTS13 binding. (A) The competition binding percentage of the VWF A2 domain variants at saturating concentrations (500 nM) to ADAMTS13 is displayed. Results are means ± SEM of at least 3 independent experiments (the N1515Q/N1574Q/Y1544D variant is labeled as Y1544D/NQ/NQ). (B) The calculated mean Tm was plotted against the mean solution binding of 500 nM VWF A2 to 30nM iADAMTS13 for the variant and condition matched VWF A2 domain variants. Example data points are indicated on the graph, and the Spearman coefficient was calculated using linear regression (GraphPad).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported by a British Heart Foundation studentship (grant FS/14/21/30733).
Authorship
Contribution: C.J.L. and D.A.L. designed the research, analyzed the results, and wrote the manuscript; and C.J.L. performed the experiments.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: David A. Lane, Centre for Haematology, Imperial College London, Room 5.S5, Commonwealth Building, Hammersmith Hospital Campus, Du Cane Rd, London W12 0NN, United Kingdom; e-mail: d.lane@imperial.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal