Key Points
With a few exceptions, the histologic and cytologic characteristics of myelodysplasia are similar in humans and mice.
As in humans, MDS and MDS/MPN are distinct diseases in mice; mouse models of these diseases can serve as useful research tools.
Abstract
Much-needed attention has been given of late to diseases specifically associated with an expanding elderly population. Myelodysplastic syndrome (MDS), a hematopoietic stem cell-based blood disease, is one of these. The lack of clear understanding of the molecular mechanisms underlying the pathogenesis of this disease has hampered the development of efficacious therapies, especially in the presence of comorbidities. Mouse models could potentially provide new insights into this disease, although primary human MDS cells grow poorly in xenografted mice. This makes genetically engineered murine models a more attractive proposition, although this approach is not without complications. In particular, it is unclear if or how myelodysplasia (abnormal blood cell morphology), a key MDS feature in humans, presents in murine cells. Here, we evaluate the histopathologic features of wild-type mice and 23 mouse models with verified myelodysplasia. We find that certain features indicative of myelodysplasia in humans, such as Howell-Jolly bodies and low neutrophilic granularity, are commonplace in healthy mice, whereas other features are similarly abnormal in humans and mice. Quantitative hematopoietic parameters, such as blood cell counts, are required to distinguish between MDS and related diseases. We provide data that mouse models of MDS can be genetically engineered and faithfully recapitulate human disease.
Introduction
Myelodysplastic syndrome (MDS) represents a heterogeneous group of hematopoietic stem cell (HSC)-based disorders,1-3 characterized by peripheral blood (PB) cytopenias of ≥1 lineage, bone marrow (BM) hypercellularity, and cytologic dysplasia.4 The term myelodysplasia encompasses all morphologic abnormalities in the affected myeloid lineage(s); its presence is a key distinguishing feature for the diagnosis of MDS. With disease progression, the hematopoietic system shows signs of genomic instability,5,6 with transformation to acute myeloid leukemia (AML) in ∼30% of patients.7
Two large, gene-focused sequencing studies evaluated MDS patient samples for mutations in >100 known cancer genes6,8 and showed that 74% to 90% of cases harbor ≥1 known oncogenic mutation in their blood cells. Both studies grouped the mutated genes according to their cellular function and found that RNA splicing was the most commonly targeted biologic process. This observation distinguishes MDS from AML, where mutations in RNA splicing genes have only been detected in a small subset of patients.9 Furthermore, it was found that genes involved in RNA splicing or DNA methylation were mutated early in the disease etiology, whereas mutations in genes essential for chromatin remodeling and cell signaling are acquired at later stages. Haferlach and colleagues8 further demonstrated that the status of a subset of 14 genes showed stand-alone, reproducible prognostic value. These studies have thus provided a molecular profile of the heterogeneous nature of MDS that will facilitate delineation of disease mechanisms and development of therapies. A major hurdle toward this goal is the apparent lack of bona fide mouse models.
Multiple attempts to create xenograft models with human MDS cells have not been successful in generating myelodysplastic features.10-17 The alternative to using xenograft mouse models is to use genetically engineered mice. However, their usefulness has been questioned because (1) unlike humans, mice do not spontaneously develop MDS as they age; (2) models engineered to develop MDS do not reflect the variety of features displayed in humans with MDS; (3) some models with genetic perturbations found in humans with MDS do not develop MDS; and (4) the criteria for diagnosing myelodysplasia in mice are not well established.18 With respect to the first concern, humans develop MDS at a frequency of ∼1/30 000 between the ages of 55 and 59 years and at ∼1/2000 at >85 years of age.19 Extrapolating these ages to laboratory mice, the detection of 1 mouse with MDS would require maintaining 30 000 mice for ∼20 months (the approximate equivalent age of 60 years in humans) or 2000 mice for ∼33 months, respectively. This would be a rather costly endeavor, and no such studies have been undertaken to our knowledge. As for the second and third concerns, mouse modeling of human disease often involves the absence, mutation, or overexpression of 1 or 2 gene products. They rarely involve a combination of >2 genetic aberrations, which is often observed in patients. Compared with the genomic complexity of human MDS,6,8 it is not surprising that some mouse models only partially recapitulate key phenotypic features. Selective cross-breeding and detailed analysis of incomplete or failed models can nevertheless help paint a more complete picture of MDS disease processes and of the interactions between genetic lesions. For example, mutations in SF3B1 are most prevalent in patients with refractory anemia with ring sideroblasts.9 In this disease entity, by definition, 15% or more of the erythroid progenitors are ring sideroblasts. However, in Sf3b1+/− mice, the presence of ring sideroblasts is either rare or nonexistent, and there is little evidence of myelodysplasia.20-22 This could be considered a failed mouse model; however, the mouse model results in reduced Sf3b1 expression levels, whereas the SF3B1 substitution mutations occurring in humans might instead confer a gain of function or encode a dominant-negative protein. Alternatively, this result might simply indicate that additional coexisting mutations are required for the development of refractory anemia with ring sideroblasts.
The fourth concern, the lack of clear diagnostic guidelines in mice, is one focus of this review. The identification of dysplastic hematopoietic cells is not trivial and requires an experienced pathologist with specialization in hematopathology. It is even more challenging in mice because serial BM sampling, to identify morphologic changes over time, is extremely difficult. It is our experience that PB myelodysplasia occurs only late in the disease history, and thus serial sampling of PB might only be helpful in recognizing a developing cytopenia. In addition to these practical difficulties, there are currently few well-established morphologic criteria for the diagnosis of dysplasia in mice.18 If mouse models are to play an informative role in MDS research, it is essential that scientists are able to make the diagnosis of MDS with confidence. This review is an effort to develop guidelines to facilitate diagnosis by expanding on the current criteria for murine myelodysplasia, as defined by the Mouse Models of Human Cancer Consortium.18 We begin by discussing the cytologic and histologic features of normal hematopoietic tissues in humans and mice, with an emphasis on the differences between them. We then discuss the diagnostic criteria of MDS in both species. Finally, we briefly discuss existing mouse models for which myelodysplastic features could be retrospectively evaluated from the images from published reports. We refer the reader who is interested in other aspects of MDS mouse models to several excellent reviews.23-25
Comparative cytologic and histologic analysis of normal human and mouse hematopoiesis
Histology
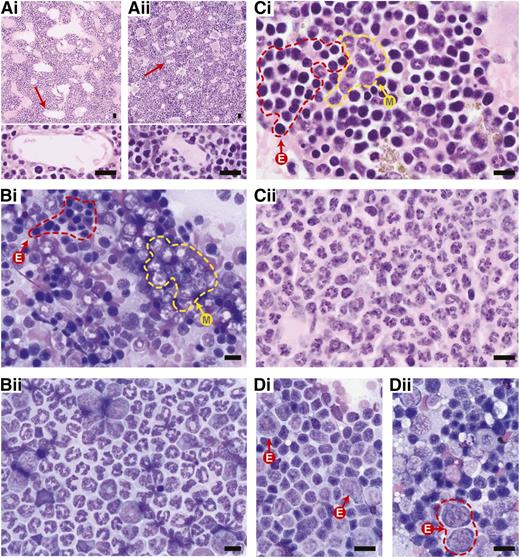
In humans, medullary fat increases with age, from ∼0% in neonates to 70% to 80% in the very old. BM hypercellularity in humans is the relative replacement of medullary fat by hematopoietic cells. In contrast, the murine medullary cavity has a more prominent network of endothelium-lined sinusoidal vascular spaces and contains significantly less fat,26,27 although this depends somewhat on what bone is being investigated.28 Because adipocytes greatly affect hematopoietic cell production,28 it is essential to use the same bones for comparison of cellular composition. The presence of flattened, channel-like sinusoids, resulting from the excess of marrow cells is an indication of hypercellularity29 (compare Figure 1, Ai with Aii). However, the best quantification of murine BM cellularity is a simple cell count from a femoral flush, normalized for weight.30 Mitotic figures, another measure of proliferation, represent <1% of all BM cells in both humans and mice.31
Representative histology of normal and aberrant hematopoiesis in mice. Images shown are either from wild-type C57Bl/6 mice (indicated by i) or from C57Bl/6 Crebbp+/− mice with MDS/MPN (indicated by ii), as examples of abnormal hematopoietic histology. (A) Compared with (Ai) H&E-stained wild-type BM, (Aii) hypercellular Crebbp+/− BM demonstrates marked compression and flattening of sinusoidal channels (red arrows and a high magnification of the indicated sinusoidal channels at the bottom) in the medullary cavity. (B-C) Giemsa-stained BM touch preparation and tissue section, respectively, show a myeloid:erythroid (M:E) ratio close to 1.5:1 in (Bi-Ci) wild-type, whereas (Bii-Cii) Crebbp+/− BM is dominated by mature segmented granulocytes). The yellow dashed line demarks areas of myelopoiesis (M); the red dashed line areas of erythropoiesis (E). (D) A Giemsa-stained wild-type spleen touch preparation contains mostly (Di) mature lymphocytes and occasional erythroid precursors (Di, arrows). In contrast, (Dii) an enlarged Crebbp+/− spleen shows extramedullary hematopoiesis with erythroid precursors (red dashed lines) and scattered granulocyte precursors with a few interspersed lymphocytes. All images in this review were produced at room temperature, using an Olympus BX51 microscope and a DP72 camera (Olympus, Center Valley, PA). Cellsens digital imaging software v.1.3 (www.olympusamerica.com) was used to capture the images. Magnification: (top panels of A) ×10; (B) ×40; and (bottom panels of A,C,D) ×60. Scale bars, 10 μm.
Representative histology of normal and aberrant hematopoiesis in mice. Images shown are either from wild-type C57Bl/6 mice (indicated by i) or from C57Bl/6 Crebbp+/− mice with MDS/MPN (indicated by ii), as examples of abnormal hematopoietic histology. (A) Compared with (Ai) H&E-stained wild-type BM, (Aii) hypercellular Crebbp+/− BM demonstrates marked compression and flattening of sinusoidal channels (red arrows and a high magnification of the indicated sinusoidal channels at the bottom) in the medullary cavity. (B-C) Giemsa-stained BM touch preparation and tissue section, respectively, show a myeloid:erythroid (M:E) ratio close to 1.5:1 in (Bi-Ci) wild-type, whereas (Bii-Cii) Crebbp+/− BM is dominated by mature segmented granulocytes). The yellow dashed line demarks areas of myelopoiesis (M); the red dashed line areas of erythropoiesis (E). (D) A Giemsa-stained wild-type spleen touch preparation contains mostly (Di) mature lymphocytes and occasional erythroid precursors (Di, arrows). In contrast, (Dii) an enlarged Crebbp+/− spleen shows extramedullary hematopoiesis with erythroid precursors (red dashed lines) and scattered granulocyte precursors with a few interspersed lymphocytes. All images in this review were produced at room temperature, using an Olympus BX51 microscope and a DP72 camera (Olympus, Center Valley, PA). Cellsens digital imaging software v.1.3 (www.olympusamerica.com) was used to capture the images. Magnification: (top panels of A) ×10; (B) ×40; and (bottom panels of A,C,D) ×60. Scale bars, 10 μm.
The cellular composition of mouse BM is distinct from humans in that it contains fewer granulocytes but more erythrocytes, monocytes, lymphocytes, and plasma cells. The myeloid-to-erythroid (M:E) ratio in normal mouse BM ranges from 0.8 to 2.5:1 (Figure 1B-C) and 1.6 to 5.4:1 in humans.32,33 The murine spleen is also different. Under normal physiologic conditions, the spleen in humans is not a site of primary hematopoiesis, whereas a low level of extramedullary hematopoiesis in the red pulp is always present in mice. This is evidenced by scattered erythroid progenitors (Figure 1D) and/or megakaryocytes throughout the clusters of lymphocytes, without disrupting the normal architecture of the spleen.
Cytology
Mature red cells in the mouse are smaller than their human equivalent, with mean cell diameters of ∼5.5 and 7 to 8 μm, respectively.34 Reticulocytes are abundant in PB smears from mice, comprising 7% to 8% of total erythrocytes in young mice to 2% to 4% in adult mice, vs 1% in humans.32 Reticulocytes are slightly larger than mature red blood cells and appear more basophilic with Wright-Giemsa staining due to more abundant RNA within them. Therefore, mild anisocytosis (ie, red cells of unequal size) and polychromasia (where red cells vary in their staining with Wright-Giemsa) are a common finding in normal mouse PB smears.32 Howell-Jolly bodies are considered pathologic in humans but can be present in normal mouse smears (usually <1% of cells).29,32
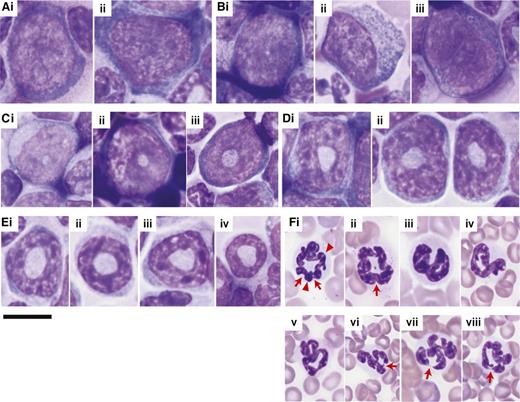
In older adult humans, neutrophils represent 40% to 70% of all white blood cells (WBCs) in the periphery. In adult mice, this proportion is only 5% to 20%. At the myeloblast stage (ie, the most immature, morphologically identifiable cell in the granulocytic lineage), the nucleus is oval, eccentrically, or peripherally placed, with fine chromatin and distinct nucleoli (Figure 2Ai-Aii). In humans, maturation changes the shape of the nucleus35 : from an oval in immature cells, it becomes flattened, progressively indenting to finally assume a multilobular or segmentation form in mature neutrophils. In mice, maturation of the granulocytic lineage starts with a small, central, nuclear clearing, which first appears at the promyelocyte stage (Figure 2Bi-Biii) and then progresses through a doughnut- or ring-shaped nuclear stage in myelocytes (Figure 2Ci-Ciii) and metamyelocytes (Figure 2Di-Dii), to become a thinner ring-like form in band cells (Figure 2Ei-Eiv). To classify a ringed cell as a neutrophil, the diameter of the center of the ring should be greater than 50% of the diameter of the nucleus.18 The nuclei of mature murine neutrophils in the PB are most often curled/ringed (Figure 2Fi-Fii) or twisted (Figure 2Fiii-Fv) but can be fully segmented (Figure 2Fvi-Fviii). Based on our experience, a normal murine segmented neutrophil contains up to 4 nuclear segments or lobes. If the number of segments is >4, it can be classified as hypersegmented. Counting segments of murine neutrophils is challenging due to the peculiar shape of the nuclei in the nonsegmented neutrophils. In these cells, the nuclei may be twisted or folded, ring-like structures, with some areas slightly bent in upon themselves. These folded regions can be mistaken for segments (Figure 2Fi-Fii, arrowheads). The key to distinguishing folding from segmentation is that definite segments are completely separated by thin, thread-like filaments of chromatin (Figure 2Fvi-Fviii, arrows) whereas a thick nuclear material connects the margins of 2 adjacent folded areas (Figure 2Fi, arrowheads). In comparison with human neutrophils, the cytoplasm of mouse neutrophils is generally plentiful and pale, and the cytoplasmic granules are fine, dust-like, and difficult to identify by Wright-Giemsa staining29 (Figure 2Fi-Fviii). This is true even in promyelocytes29 (Figure 2B), a stage at which the azurophilic granules are prominent in human cells (see figure 1 of Mufti et al35 ).
Morphologic characteristics of the different stages of granulocytic maturation in wild-type mice. Images are of Giemsa-stained (A-E) BM touch preparations and (F) PB smears. The series of pictures from A to F represent the granulocytic differentiation process from the most immature precursors to the most mature form. (Ai-Aii) Myeloblasts have oval nuclei with fine chromatin and distinct nucleoli. Some (Bi-Bii) promyelocytes start to show a small, central clearing in the nucleus that indicates the beginning of the maturation process. The granules in murine promyelocytes are difficult to discern in comparison with their human counterpart. The nuclear clearing enlarges with increasing differentiation, transforming the nucleus to ring-like structures in (Ci-Ciii) myelocytes, (Di-Dii) metamyelocytes, and (Ei-Eiv) band cells, where the string-like form is thinnest. The nuclei of mature neutrophils are most often (Fi-Fii) curled/ringed or (Fiii-Fv) twisted but can also be (Fvi-Fviii) fully segmented. Arrows point to filaments of chromatin separating nuclear segments; arrowheads point to nuclear folds. Of note, myelocytes and metamyelocytes may be difficult to distinguish from immature monocytes because all may have ring-shaped nuclei and a pale blue cytoplasm. Magnification: ×60. Scale bars, 10 μm for all images.
Morphologic characteristics of the different stages of granulocytic maturation in wild-type mice. Images are of Giemsa-stained (A-E) BM touch preparations and (F) PB smears. The series of pictures from A to F represent the granulocytic differentiation process from the most immature precursors to the most mature form. (Ai-Aii) Myeloblasts have oval nuclei with fine chromatin and distinct nucleoli. Some (Bi-Bii) promyelocytes start to show a small, central clearing in the nucleus that indicates the beginning of the maturation process. The granules in murine promyelocytes are difficult to discern in comparison with their human counterpart. The nuclear clearing enlarges with increasing differentiation, transforming the nucleus to ring-like structures in (Ci-Ciii) myelocytes, (Di-Dii) metamyelocytes, and (Ei-Eiv) band cells, where the string-like form is thinnest. The nuclei of mature neutrophils are most often (Fi-Fii) curled/ringed or (Fiii-Fv) twisted but can also be (Fvi-Fviii) fully segmented. Arrows point to filaments of chromatin separating nuclear segments; arrowheads point to nuclear folds. Of note, myelocytes and metamyelocytes may be difficult to distinguish from immature monocytes because all may have ring-shaped nuclei and a pale blue cytoplasm. Magnification: ×60. Scale bars, 10 μm for all images.
The morphology of eosinophils differ between humans and mice.36 In the former, most eosinophils are bi-lobulated, whereas murine eosinophils are ring formed, similar to mature neutrophils. The less compact chromatin structure of murine eosinophils distinguishes them from neutrophils.
The majority (80-90%) of peripheral WBCs in older adult mice are mature lymphocytes, in contrast to their human counterparts, where lymphocytes comprise only 20% to 40%. The morphology of mature murine lymphocytes, monocytes, and basophils is similar to that of humans. Irrespective of the strain, the laboratory mouse has a very high platelet count compared with humans: 1013 to 1633 × 109/L vs 150 to 400 × 109/L, respectively.32,37 This high platelet number in the PB may account for the presence of platelet clumps, especially after a difficult sampling. Finally, murine WBCs seem more fragile than their human counterparts and thus are more easily damaged during smear preparation. The presence of basket cells (or smudge cells) in smears, a characteristic sign of chronic lymphocytic leukemia in humans, usually simply indicates poor slide preparation in mice.32
In summary, several histologic and cytologic features, which in humans would be diagnostic for hematologic diseases, are part of normal murine hematopoiesis. In particular, the presence of Howell-Jolly bodies in the PB and the relative lack of granules in cells of the granulocytic lineage cannot be interpreted as myelodysplasia in mice.
Diagnostic criteria for MDS in humans and mice
The criteria for diagnosing MDS in humans and mice are presented in Table 1. To evaluate these criteria requires a combination of clinical course, complete blood counts, and morphologic and immunophenotypic data. In both species, MDS may be diagnosed when there is PB cytopenia and evidence of dysplasia in ≥1 of the 3 myeloid lineages (necessary criteria). For humans, the presence of ≥10% dysplastic cells within a particular lineage is considered abnormal, although the suitability of this frequency as a diagnostic threshold is still under debate.38 Knowledge about what constitutes the minimal criteria for dysplasia in mice is currently lacking because the proportion of dysplastic cells is rarely reported in mouse studies. To improve the quality of MDS mouse models, we recommend that, in future studies, the proportion of dysplastic cells is reported. Table 2 presents the cytologic and histologic characteristics that, according to the World Health Organization39,40 and the Mouse Models of Human Cancer Consortium,18 constitute dysplasia in the respective species. Molecular signs of clonal disease and/or reduced numbers of colony-forming BM progenitors may facilitate diagnosing MDS (supportive criteria). However, these 2 features can also be found in healthy individuals and are therefore by themselves insufficient to confer a diagnosis of MDS. In addition to the necessary and supportive criteria, signs of leukemic transformation (ie, >20% blasts in the PB or BM) or other underlying causes of cytopenias or dysplasia must be absent (exclusion criteria). Potentially confounding entities are the MDS and myeloproliferative neoplasm (MPN) overlap syndromes (MDS/MPN).39,40 There are no prominent myeloproliferative features in human MDS, in contrast to MDS/MPN, where patients show excessive proliferation in 1 of the PB myeloid lineages (often thrombocytosis) with or without cytopenia in ≥1 of the other lineages. MDS/MPN may be accompanied by splenomegaly (ie, an enlarged spleen), but this is not an essential diagnostic feature.41,42
Diagnostic criteria for MDS in humans and mice
| . | Human* . | Mouse† . |
|---|---|---|
| Necessary criteria | ||
| Cytopenia in at least one lineage | Anemia (Hb <11 g/dL) | Anemia‡ (without leukocytosis or thrombocytosis) |
| Neutropenia (ANC <1500/µL) | Neutropenia‡ (with or without anemia or thrombocytopenia) | |
| Thrombocytopenia (PLT <1 × 105/µL) | Thrombocytopenia‡ (without leukocytosis or erythrocytosis | |
| Dyspoiesis in at least one lineage | >10% dysplastic cells in ≥1 lineage | Dyspoiesis with or without increased numbers of immature nonlymphoid cells |
| Supportive criteria | Molecular evidence of a monoclonal cell population | Molecular evidence of a monoclonal cell population |
| Persistent decrease in colony-forming hematopoietic progenitors | ||
| Exclusion criteria | Other disorders that can cause cytopenia or dysplasia | >20% nonlymphoid blasts in the marrow or spleen (suggesting a nonlymphoid leukemia) |
| . | Human* . | Mouse† . |
|---|---|---|
| Necessary criteria | ||
| Cytopenia in at least one lineage | Anemia (Hb <11 g/dL) | Anemia‡ (without leukocytosis or thrombocytosis) |
| Neutropenia (ANC <1500/µL) | Neutropenia‡ (with or without anemia or thrombocytopenia) | |
| Thrombocytopenia (PLT <1 × 105/µL) | Thrombocytopenia‡ (without leukocytosis or erythrocytosis | |
| Dyspoiesis in at least one lineage | >10% dysplastic cells in ≥1 lineage | Dyspoiesis with or without increased numbers of immature nonlymphoid cells |
| Supportive criteria | Molecular evidence of a monoclonal cell population | Molecular evidence of a monoclonal cell population |
| Persistent decrease in colony-forming hematopoietic progenitors | ||
| Exclusion criteria | Other disorders that can cause cytopenia or dysplasia | >20% nonlymphoid blasts in the marrow or spleen (suggesting a nonlymphoid leukemia) |
ANC, absolute neutrophil count; cytopenia, reduced number of blood cells; cytosis, increased number of blood cells; dyspoiesis, abnormal cellular maturation; Hb, hemoglobin; PLT, platelet.
Data adapted from Kogan et al.18
Threshold numbers are not provided here because they differ for each mouse strain. For information about normal values for particular mouse strains, please see Russell and Bernstein.44
Characteristics of myelodysplasia in humans and mice
| Lineage . | Human* . | Mouse† . |
|---|---|---|
| Erythroid | Binucleated erythroid percursors | Multinucleated erythroid precursors |
| Nuclear budding, nuclear strings or internuclear bridging | Karyorrhexis | |
| Karyorrhexis | Irregular nuclear contours | |
| Cytoplasmic fraying | Ringed sideroblasts | |
| ≥15% ringed sideroblasts | Megaloblastic change with asynchrony | |
| Granulocytic | Hypersegmented nuclei | Abnormal cytoplasmic maturation |
| Hyposegmented nuclei | Abnormal nuclear maturation | |
| Pseudo-Pelger-Huet anomaly | Hypersegmentation | |
| Abnormal granulation: hypogranulation, pseudo Chediak-Higashi large granules, dimorphic granules (basophilic and eosinophilic granules) within eosinophils | ||
| Auer rods | ||
| Megakaryocytic | Hypersegmented nuclei | Strange hypersegmentation |
| Hyposegmented nuclei | Unilobulated nuclei | |
| Multiple separated nuclei | Multiple separated nuclei | |
| Micromegakaryocytes | Micromegakaryocytes | |
| Ballooning of platelets |
| Lineage . | Human* . | Mouse† . |
|---|---|---|
| Erythroid | Binucleated erythroid percursors | Multinucleated erythroid precursors |
| Nuclear budding, nuclear strings or internuclear bridging | Karyorrhexis | |
| Karyorrhexis | Irregular nuclear contours | |
| Cytoplasmic fraying | Ringed sideroblasts | |
| ≥15% ringed sideroblasts | Megaloblastic change with asynchrony | |
| Granulocytic | Hypersegmented nuclei | Abnormal cytoplasmic maturation |
| Hyposegmented nuclei | Abnormal nuclear maturation | |
| Pseudo-Pelger-Huet anomaly | Hypersegmentation | |
| Abnormal granulation: hypogranulation, pseudo Chediak-Higashi large granules, dimorphic granules (basophilic and eosinophilic granules) within eosinophils | ||
| Auer rods | ||
| Megakaryocytic | Hypersegmented nuclei | Strange hypersegmentation |
| Hyposegmented nuclei | Unilobulated nuclei | |
| Multiple separated nuclei | Multiple separated nuclei | |
| Micromegakaryocytes | Micromegakaryocytes | |
| Ballooning of platelets |
Myelodysplastic features and pitfalls illustrated
Our laboratory has extensively studied hematopoiesis in the context of hemizygosity of the CREB binding protein (Crebbp) gene, and we will use this model for illustrative purposes. Naïve Crebbp+/− mice show HSC defects,45 BM hypercellularity, splenomegaly, and myelodysplasia with hypersegmented granulocytes and pseudo-Pelger-Huet anomalies in the PB and abnormal megakaryocytes (ie, hyperlobulation and naked nuclei) in the BM.30 There is no cytopenia. Instead, these mice show PB granulocytosis and excessive myelopoiesis in marrow and spleen.30 The hematologic syndrome observed in naïve Crebbp+/− mice was therefore classified as an MDS/MPN overlap disease.40,42,46,47 The BM microenvironment contributes to the myeloproliferative component of the hematologic disease observed in Crebbp+/− mice, in part through the altered production levels of KITL and MMP9.47 In the absence of the proliferative stimulus of the Crebbp+/− stroma (eg, when wild-type mice are transplanted with Crebbp+/− Lin−Sca-1+cKit++ cells [LSKs]), recipients develop MDS, characterized by myelodysplasia in 3 lineages and leukopenia without a hyperproliferative component. Nearly half of the recipients also develop anemia with or without thrombocytopenia (T. Zhou, S. Perez, Z. Cheng, M. Kinney, L. Scott, and V. Rebel, unpublished data, March 2014). Interestingly, transplantation of nonfractionated Crebbp+/− BM results in leukemia, MDS, or MDS/MPN (T. Zhou, S. Perez, Z. Cheng, M. Kinney, L. Scott, and V. Rebel, unpublished data, March 2014).48
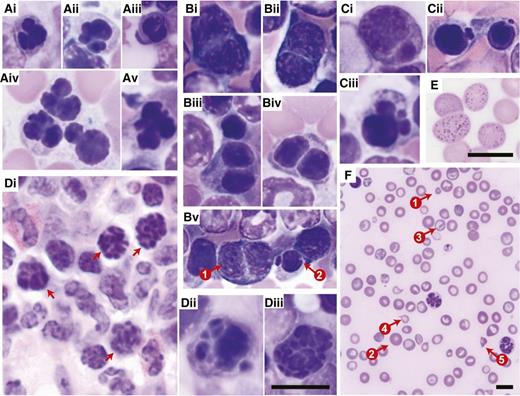
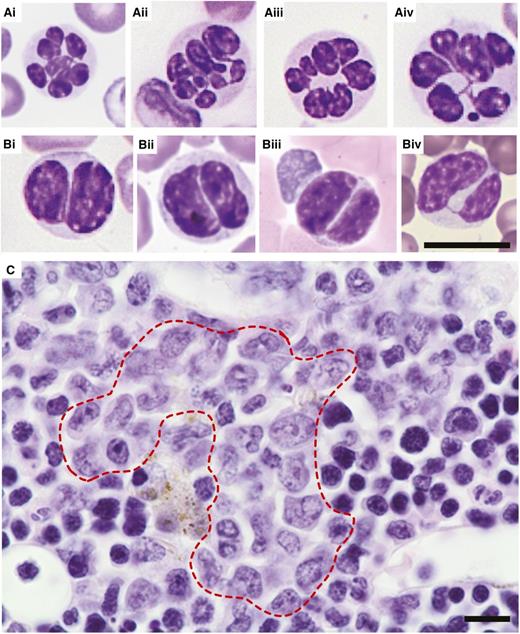
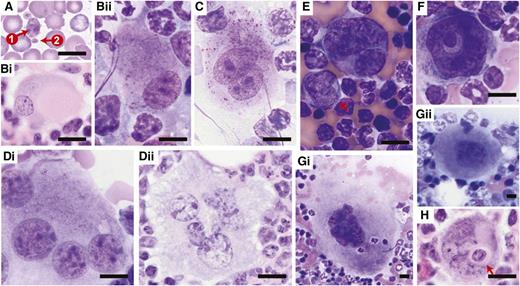
Figures 3 to 5 show the dysplastic features we typically observed in the erythroid, myeloid, and megakaryocytic lineages of the transplant models. In the erythroid lineage, myelodysplasia in the BM was evidenced by nuclear irregularity (Figure 3Ai-Av), binucleation (Figure 3Bi-Bv), nuclear budding (Figure 3Ci-Ciii), and karyorrhexis (ie, nuclear fragmentation within a dying cell; Figure Di-Diii). In the PB, abnormal erythropoiesis manifested itself by basophilic stippling (Figure 3Ei) and anisocytosis, as well as poikilocytosis (ie, the presence of abnormally shaped red cells such as target cells and teardrop cells; Figure 3F). The myeloid lineage showed hypersegmented granulocytes (Figure 4Ai-Aiv) and pseudo-Pelger-Huet anomalies in the PB (Figure 4Bi-Biv). In humans, “atypical localization of immature precursors” can be present in MDS and is often an early sign of leukemogenesis.49 This feature (Figure 4C) was noted in some of our mice with MDS. The megakaryotic lineage showed giant platelets (Figure 5A), monolobulation (Figure 5Bi-Bii), binucleation (Figure 5C), multinucleation (Figure 5Di-Dii), and micro-megakaryocytes (Figure 5E). In addition, we observed megakaryocytic abnormalities that are not considered MDS specific, but are indicative of maturational abnormalities, such as a ring-shaped nucleus (Figure 5F) and gigantic size (Figure 5Gi; compare with wild-type control cells at the same magnification shown in Figure 5Gii). We also observed megakaryocytic emperipolesis (phagocytosis) (Figure 5H), but this is not indicative of pathology.
Dysplasia in the erythroid lineage. Representative images are of (A-C,Dii-Diii) Giemsa-stained BM touch preparations, (Di) H&E-stained bone sections, and (E-F) Giemsa-stained PB smears obtained from wild-type recipients transplanted with Crebbp+/− LSKs or unfractionated BM cells. (A) Erythroid precursors with abnormal nuclear contours (lobulation). (Bi-Biv) Binucleated erythroid precursors. (Bv) Coexistence of (1) a binucleated erythroid precursor and (2) an erythroid precursor with nuclear budding. (Ci-Ciii) Erythroid precursors with nuclear budding. (Di-Diii) Karyorrhexis in erythroid precursors. (E) Basophilic stippling in peripheral red blood cells. (F) Anisopoikilocytosis, (1) microcytes, (2) macrocytes, (3) target cells, (4) tear-drop cells, and (5) red blood cell fragments. Magnification: (A-E) ×60 and (F) ×20. Scale bars, 10 μm; scale bar provided in Diii serves all images included in A to D.
Dysplasia in the erythroid lineage. Representative images are of (A-C,Dii-Diii) Giemsa-stained BM touch preparations, (Di) H&E-stained bone sections, and (E-F) Giemsa-stained PB smears obtained from wild-type recipients transplanted with Crebbp+/− LSKs or unfractionated BM cells. (A) Erythroid precursors with abnormal nuclear contours (lobulation). (Bi-Biv) Binucleated erythroid precursors. (Bv) Coexistence of (1) a binucleated erythroid precursor and (2) an erythroid precursor with nuclear budding. (Ci-Ciii) Erythroid precursors with nuclear budding. (Di-Diii) Karyorrhexis in erythroid precursors. (E) Basophilic stippling in peripheral red blood cells. (F) Anisopoikilocytosis, (1) microcytes, (2) macrocytes, (3) target cells, (4) tear-drop cells, and (5) red blood cell fragments. Magnification: (A-E) ×60 and (F) ×20. Scale bars, 10 μm; scale bar provided in Diii serves all images included in A to D.
Dysplasia in the myeloid lineage. Representative images of (A-B) Giemsa-stained PB smears and (C) H&E-stained BM sections obtained from wild-type recipients transplanted with Crebbp+/− LSKs or unfractionated BM cells. (A) Hypersegmented granulocytes. (B) Pseudo-Pelger-Huet anomalies in bilobed cells most consistent with neutrophils. (C) Atypical localization of immature precursors (red dashed line; ie, clusters of myeloid precursors present in the intertrabecular area, rather than adjacent to trabeculae or surrounding endothelial cells as is the case in wild-type hematopoiesis). Magnification: ×60. Scale bars, 10 μm; scale bar provided in Biv serves all images included in A and B.
Dysplasia in the myeloid lineage. Representative images of (A-B) Giemsa-stained PB smears and (C) H&E-stained BM sections obtained from wild-type recipients transplanted with Crebbp+/− LSKs or unfractionated BM cells. (A) Hypersegmented granulocytes. (B) Pseudo-Pelger-Huet anomalies in bilobed cells most consistent with neutrophils. (C) Atypical localization of immature precursors (red dashed line; ie, clusters of myeloid precursors present in the intertrabecular area, rather than adjacent to trabeculae or surrounding endothelial cells as is the case in wild-type hematopoiesis). Magnification: ×60. Scale bars, 10 μm; scale bar provided in Biv serves all images included in A and B.
Dysplasia in the megakaryocytic lineage. Representative images are of (A) Giemsa-stained peripheral blood smears, Giemsa-stained (Bii,C,Di,E,F,Gi,Gii) BM touch preparations, and (Bi,Dii,H) H&E-stained bone sections obtained from wild-type recipients transplanted with Crebbp+/− LSKs or unfractionated BM cells. (A) Giant platelet (arrow 1) in comparison with normal-sized platelet (arrow 2). (Bi-Bii) Megakaryocytes with eccentric, monolobated nuclei. (C) Binucleated megakaryocyte. (Di-Dii) Multinucleated megakaryocytes. (E) Micro-megakaryocyte (arrow). (F) Megakaryocyte with a ring-shaped nucleus. (Gi) A giant megakaryocyte. The size of this particular one is 10 804.9 μm2, in comparison with (Gii) a wild-type megakaryocyte of 1843.7 μm2 in size. (H) Emperipolesis of neutrophils (arrow) within megakaryocyte. Magnification: ×60. Scale bars, 10 μm. Of note, bone sections sliced at any particular level merely provide a 2-dimensional representation of the 3-dimensional BM tissue. The appearance of mono/hypolobulation may result from superficial sectioning of a deeper, well-lobulated megakaryocyte. Similarly, what seems to be multiple nuclei may actually represent different lobes of the same nucleus that are connected to each other at a deeper level tissue section. Therefore, although bone sections provide a good approach for preserving the architecture of BM, they can convey a misleading impression of cell morphology, and thus caution needs to be taken when evaluating the nuclear lobes of megakaryocytes. A more accurate evaluation can be made using BM touch preparations where complete cells are attached to the slides.
Dysplasia in the megakaryocytic lineage. Representative images are of (A) Giemsa-stained peripheral blood smears, Giemsa-stained (Bii,C,Di,E,F,Gi,Gii) BM touch preparations, and (Bi,Dii,H) H&E-stained bone sections obtained from wild-type recipients transplanted with Crebbp+/− LSKs or unfractionated BM cells. (A) Giant platelet (arrow 1) in comparison with normal-sized platelet (arrow 2). (Bi-Bii) Megakaryocytes with eccentric, monolobated nuclei. (C) Binucleated megakaryocyte. (Di-Dii) Multinucleated megakaryocytes. (E) Micro-megakaryocyte (arrow). (F) Megakaryocyte with a ring-shaped nucleus. (Gi) A giant megakaryocyte. The size of this particular one is 10 804.9 μm2, in comparison with (Gii) a wild-type megakaryocyte of 1843.7 μm2 in size. (H) Emperipolesis of neutrophils (arrow) within megakaryocyte. Magnification: ×60. Scale bars, 10 μm. Of note, bone sections sliced at any particular level merely provide a 2-dimensional representation of the 3-dimensional BM tissue. The appearance of mono/hypolobulation may result from superficial sectioning of a deeper, well-lobulated megakaryocyte. Similarly, what seems to be multiple nuclei may actually represent different lobes of the same nucleus that are connected to each other at a deeper level tissue section. Therefore, although bone sections provide a good approach for preserving the architecture of BM, they can convey a misleading impression of cell morphology, and thus caution needs to be taken when evaluating the nuclear lobes of megakaryocytes. A more accurate evaluation can be made using BM touch preparations where complete cells are attached to the slides.
The Crebbp+/− mouse models illustrates 2 important aspects of mouse myelodysplasia: first, dysplastic features in mice closely resemble those described for humans with MDS; second, the presence of myelodysplasia in mice is not per se sufficient for the diagnosis of MDS, and, as with humans, additional information such as blood counts and organomegaly are necessary to rule out MDS/MPN.
Mouse models with dysplasia in one or more hematopoietic lineages
In addition to the Crebbp+/− mouse models, we found 21 others for which we could confirm myelodysplastic features from published images. The genetic manipulations used to generate these models include gene deletion (Asxl1, CD74-Nid67, Bap1, Dicer, Dnmt3a, Map3k7/Tak, miR145/146a, and Npm1),50-58 overexpression (mutated Asxl1, Evi1, Nup98-Hox13D, mutated Runx1, human [h]SALL4b, hS100A9, Bcl2 + mutated NRAS and Traf6),1,56,59-65 and mutation (CyclinE and Polg).66,67 All but 4 of these genes have corresponding alterations in human MDS (or MDS/MPN) patients (Table 3). The link between human MDS and those 4 genes (Cyclin E, Dicer, Polg, and hSALL4B) may not exist or may simply remain to be discovered.
Genetic targets in mouse models with myelodysplasia and their implication in human MDS or MDS/MPN
| Gene symbol . | Frequency (%)* . | Predominant genetic aberration . | Reference . | |
|---|---|---|---|---|
| MDS . | MDS/MPN . | |||
| ASXL1 | 11-20.7 | 33-49† | Mutation | 68,,,,,-74 |
| BAP1 | 3‡ | Truncating mutation | 52 | |
| CD74-NID67 | + | Deleted in 5q- syndrome§ | ||
| CREBBP | + | 2 cases | Translocation¶ | 75,-77 |
| <2 | Mutation | 6 | ||
| CCNE1 (CYCLIN E) | ||||
| DICER | ||||
| DNMT3A | 0-13.5 | 0-6.8 | Mutation | 78 |
| EVI1 | + | Translocation | 79 | |
| 26-29 | Overexpressed in blood cells from MDS patients | 80,81 | ||
| MAP3K7 (TAK1) | 22† | Gene deletion | 55 | |
| miR145/146a | + | Significantly reduced expression in blood cells from patients with 5q- syndrome | 56 | |
| NPM1 | 0-5.8 | 0-14.3 | Mutation | 70,74,82,,,-86 |
| NRAS | 5.7-6.3 | 17-19 | G12D mutation# | 87,-89 |
| NUP98-HOXD13 | 1 case | Translocation | 90 | |
| POLG | ||||
| RUNX1 | 12-13.8 | 37† | D171N, S291fs# | 87,91 |
| SALL4b | ||||
| S100A9 | + | Increased number of myeloid cells expressing S100A9 are found in MDS patients | 64 | |
| TRAF6 | + | Possibly increased in blood cells of patients with 5q- syndrome | 92 | |
| Gene symbol . | Frequency (%)* . | Predominant genetic aberration . | Reference . | |
|---|---|---|---|---|
| MDS . | MDS/MPN . | |||
| ASXL1 | 11-20.7 | 33-49† | Mutation | 68,,,,,-74 |
| BAP1 | 3‡ | Truncating mutation | 52 | |
| CD74-NID67 | + | Deleted in 5q- syndrome§ | ||
| CREBBP | + | 2 cases | Translocation¶ | 75,-77 |
| <2 | Mutation | 6 | ||
| CCNE1 (CYCLIN E) | ||||
| DICER | ||||
| DNMT3A | 0-13.5 | 0-6.8 | Mutation | 78 |
| EVI1 | + | Translocation | 79 | |
| 26-29 | Overexpressed in blood cells from MDS patients | 80,81 | ||
| MAP3K7 (TAK1) | 22† | Gene deletion | 55 | |
| miR145/146a | + | Significantly reduced expression in blood cells from patients with 5q- syndrome | 56 | |
| NPM1 | 0-5.8 | 0-14.3 | Mutation | 70,74,82,,,-86 |
| NRAS | 5.7-6.3 | 17-19 | G12D mutation# | 87,-89 |
| NUP98-HOXD13 | 1 case | Translocation | 90 | |
| POLG | ||||
| RUNX1 | 12-13.8 | 37† | D171N, S291fs# | 87,91 |
| SALL4b | ||||
| S100A9 | + | Increased number of myeloid cells expressing S100A9 are found in MDS patients | 64 | |
| TRAF6 | + | Possibly increased in blood cells of patients with 5q- syndrome | 92 | |
Presented are the frequencies of sequence abnormalities observed in human MDS or MDS/ MPN patients. A + indicates that a perturbation was found but no frequency was reported.
In patients with chronic myelomonocytic leukemia.
One of 32 patients tested showed a mutation in BAP1.
Five percent to 10% of MDS patients show deletion of 5q.93,94 The region deleted in the mouse model represents the smallest commonly deleted region in an analysis of 16 5q- MDS patients.95
Crebbp translocations mostly involve therapy-related hematopoietic malignancies.
Deletion of the Dicer gene occurred in the nonhematopoietic osteoprogenitor compartment and was sufficient for development of myelodysplasia and secondary leukemia.53 Although the BM microenvironment has been recognized in human MDS development,97 very little effort has been made to date to identify driver mutations in the nonhematopoietic marrow cells of MDS patients. This may explain why mutations or other aberrations in DICER have yet to be documented in MDS or any other hematopoietic malignancies.
Two other MDS mouse models are worth mentioning in the context of microenvironmental effects on MDS development: First, overexpression of hS100A9 in a committed myeloid cell population (thus not in HSCs) was sufficient to cause MDS development.64 Second, PolgD257A animals developed fatal megaloblastic anemia; however, when PolgD257A/D257A BM cells were transplanted into wild-type recipients, the onset of disease was earlier than in the donor mouse, and a significant thrombocytosis started to manifest 1 month after transplantation.67 The authors concluded that the age of donor HSCs dictates the course of the disease, consistent with the possibility of accumulating mitochondrial damage. However, an alternative explanation may be that the outcome depends on the microenvironmental context in which HSCs reside.
Clinical features
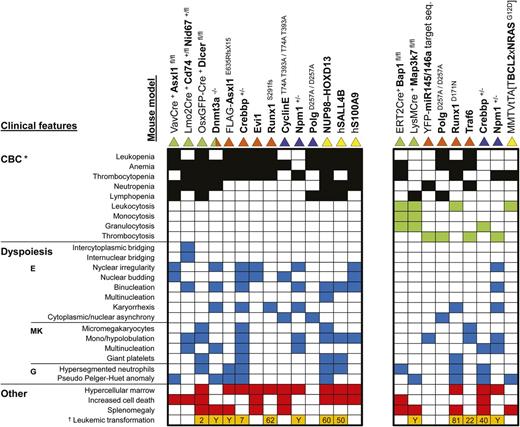
It is important to note that some mouse models with myelodysplasia also display PB cytosis. In humans, this would warrant the diagnosis MDS/MPN overlap disease.40,41,46 In keeping with this distinction in humans, we divided the models into 2 categories: those with cytopenia only and those that showed significant peripheral cytosis. Based on this distinction, 14 mouse models could be classified as MDS models (Figure 6, left block) and 9 as MDS/MPN (Figure 6, right block).
Critical disease features in mice with myelodysplasia. Block plot showing clinical features (rows) present in the different mouse models (columns). The presence of dysplastic characteristics in the myeloid lineages identified to the left of the figure is indicated by blue squares. In addition to myelodysplasia, the mouse models in the right block show cytosis in one or more lineages (green squares), which in most cases is accompanied by cytopenia in a different lineage (black squares). The left block includes models that show only cytopenia (no cytosis). The red squares indicate the presence of a hypercellular marrow, increased cell death, or splenomegaly. Orange squares signify that leukemic transformation occurs in these animals. White squares indicate that the respective feature was absent or not reported for the model. The color(s) of the triangle indicate(s) the type of mouse model presented in the respective column: green indicates a conditional knockout model; orange a BM transplantation model with wild-type mice as recipients; blue, a knockout or knock-in model; yellow, a transgenic model. One of the 45 recipients transplanted with Dnmt3a−/− HSCs developed chronic myelomonocytic leukemia, which is classified as an MDS/MPN overlap disease. Because it was only 1 mouse, this model was not included in the MDS/MPN category. *Peripheral blood cytopenia(s) in 1 of the 3 myeloid lineages is a requirement for the diagnosis of MDS. Leukopenia only signals that the total number of leukocytes is significantly lower than in control mice. However, it does not distinguish between lymphocytes, neutrophils, and monocytes. Information about the size of these subpopulations is essential to make the correct diagnosis. In addition, leukopenia that is only based on a lymphopenia does not fulfill the requirement for the diagnosis of MDS. †Numbers in the respective boxes represent the percentages that develop leukemia, whereas a “Y” denotes the fact that leukemia progression was reported but the proportion of animals was not clear.
Critical disease features in mice with myelodysplasia. Block plot showing clinical features (rows) present in the different mouse models (columns). The presence of dysplastic characteristics in the myeloid lineages identified to the left of the figure is indicated by blue squares. In addition to myelodysplasia, the mouse models in the right block show cytosis in one or more lineages (green squares), which in most cases is accompanied by cytopenia in a different lineage (black squares). The left block includes models that show only cytopenia (no cytosis). The red squares indicate the presence of a hypercellular marrow, increased cell death, or splenomegaly. Orange squares signify that leukemic transformation occurs in these animals. White squares indicate that the respective feature was absent or not reported for the model. The color(s) of the triangle indicate(s) the type of mouse model presented in the respective column: green indicates a conditional knockout model; orange a BM transplantation model with wild-type mice as recipients; blue, a knockout or knock-in model; yellow, a transgenic model. One of the 45 recipients transplanted with Dnmt3a−/− HSCs developed chronic myelomonocytic leukemia, which is classified as an MDS/MPN overlap disease. Because it was only 1 mouse, this model was not included in the MDS/MPN category. *Peripheral blood cytopenia(s) in 1 of the 3 myeloid lineages is a requirement for the diagnosis of MDS. Leukopenia only signals that the total number of leukocytes is significantly lower than in control mice. However, it does not distinguish between lymphocytes, neutrophils, and monocytes. Information about the size of these subpopulations is essential to make the correct diagnosis. In addition, leukopenia that is only based on a lymphopenia does not fulfill the requirement for the diagnosis of MDS. †Numbers in the respective boxes represent the percentages that develop leukemia, whereas a “Y” denotes the fact that leukemia progression was reported but the proportion of animals was not clear.
MDS models
Consistent with the clinical presentation of human MDS, the features that lead to the diagnosis of MDS in mice present a heterogeneous picture (Figure 6, left block). The majority of models display cytopenia in 2 or 3 lineages (79%), with the erythroid lineage being most frequently affected (93%). In human MDS, erythropoiesis is also more commonly affected than the other hematopoietic processes.98 The degree and types of myelodysplasia vary: 5 of 14 models show myelodysplasia in 1 lineage, whereas the remaining 9 have involvement of 2 or 3 lineages. Dysplasia involves the erythroid lineage in 86% of the models, whereas the megakaryocytic and granulocytic lineages are affected in 57% and 50%, respectively. Ten models have hypercellular marrows but only 5 present with splenomegaly. Conditional deletion of Asxl199 results in MDS with a hypocellular marrow, which, although not typical, does occur in human MDS.100 Interestingly, when Tet2 was deleted in addition to Asxl1, mice developed MDS with a hypercellular marrow and normocellular spleen.99 As with humans, murine MDS models do not always develop leukemia (6 of 12); those that do show frequencies of transformation between 2% and 62%.
MDS/MPN overlap disease models
Mouse models presented on the right in Figure 6 differ from those on the left by the presence of peripheral cytosis. In addition, they show fewer myelodysplastic features: 67% (6 of 9) present with dysplasia in a single lineage and the erythroid lineage is only affected in 33% of the models. Splenomegaly was more frequent (5 of 9 vs 5 of 14) in the MDS/MPN models than in the MDS ones, whereas hypercellular marrow (3 of 9 vs 10 of 14) and increased cell death (2 of 9 vs 8 of 14) were both less commonly seen. As with the MDS models, transformation to leukemia was variable, both in occurrence (56%) and in penetrance (22-81%).
Models that give rise to more than one disease entity
Npm1 heterozygosity results in myelodysplasia with high penetrance (80%) between 6 and 18 months of age.57 It is characterized by dysplasia and other abnormalities in the erythroid lineage and in the megakaryocytic lineage (Figure 6). PB platelet analysis revealed a wide distribution of values: of the 9 animals described, 2 had a normal platelet count, whereas the others showed either thrombocytopenia or thrombocytosis, in roughly equal proportions. Thus, Npm1+/− mice develop both MDS and MDS/MPN. Approximately 16% of Npm1+/− mice developed a hematopoietic malignancy, mostly of myeloid origin.58 Overall, this mouse model shows a heterogeneity in disease presentation that is reminiscent of humans with myelodysplasia. Long-term follow-up of recipients transplanted with Dnmt3a−/− HSCs revealed a variety of hematologic diseases, as predicted by the widespread occurrence of DNMT3A mutations in human hematopoietic malignancies.78 Of the mice with an unambiguous diagnosis of a hematopoietic malignancy, 82% developed a myeloid neoplasm and 19% a lymphoid neoplasm.54 MDS occurred in 38% of the cases and was characterized by ≥1 PB cytopenia and tri-lineage myelodysplasia (Figure 6).
Concluding remarks
Many of the genes thought to be important for MDS disease formation in humans have been genetically manipulated in mice.23-25 These models have provided essential insights into the role of these genes in maintaining the functional and/or genomic integrity of HSCs (believed to be the cells of origin in MDS); however, not all of these models show MDS development. This has led to some skepticism about the use of mouse models in understanding MDS. It is becoming clear that MDS is a disease of genomic instability, defined as the continuous accumulation of chromosomal, oligonucleotide, and base pair abnormalities.101 Moreover, several studies have revealed the importance of cooperative mutations in MDS development.6,8 As a consequence, it is likely that some mouse models failed to produce MDS because a single gene perturbation was not an adequate trigger. We nevertheless identified 23 mouse models that clearly displayed myelodysplasia, a key feature of MDS. Detailed analysis of these models revealed that, with a few important exceptions, very similar histopathologic features could be used to characterize myelodysplasia in mice and in humans (Figures 3-6; Table 2). Most models belonged to 1 of 2 groups: those that showed myelodysplasia and PB cytopenia but without evidence of PB hyperproliferation, and those where myelodysplasia was accompanied by PB cytosis (hyperproliferation) in 1 of the 3 myeloid lineages (Figure 6). According to the World Health Organization, these sets of features correspond to MDS and MDS/MPN overlap disease, respectively39-41 (although 5q- MDS occasionally presents with thrombocytosis). Whether and how these syndromes are related is a clinically relevant question, which can possibly be addressed by further comparative analysis of Npm1+/− mice with either disease or mouse models with the same gene perturbation but different outcome in a transplant setting, such as the Crebbp+/− and PolgD257A/D257A mouse models. Until we know the relationship between these 2 diseases, MDS and MDS/MPN overlap disease mouse models should not be used interchangeably.
It is our conclusion that MDS and MDS/MPN in mice are sufficiently similar to their human counterparts for genetically engineered mouse models of these diseases to serve as useful research tools. They would be particularly valuable in studies difficult or impossible to perform in humans or in xenograft models such as pinpointing the initiating events of MDS in a predisease state. The search for biomarkers predictive of increased risk of developing MDS or of progressing to leukemia would also benefit from these models, as would drug development studies.
Acknowledgments
The authors thank Dr Madeleine Lemieux for editing the manuscript (http://bioinfo.ca/) and David Rodriguez for help with preparing the images for publication.
This publication was supported by funding from the Greehey Children's Cancer Research Institute (to V.I.R.), the National Institutes of Health, National Institute of Environmental Health Sciences grant 1RO1ES022054 (to V.I.R.), and the University of Texas System's Graduate Programs Initiative that funded the Translational Science Training Across Disciplines program at the University of Texas Health Science Center at San Antonio (T.Z.). T.Z. was also supported by a Hyundai Hope Grant Award.
Authorship
Contribution: T.Z. captured all histologic photographs and wrote the manuscript; M.C.K. provided guidance on the pathologic aspects of this article; L.M.S. provided guidance on MPN-related issues; S.S.Z. provided guidance on human MDS and MDS/MPN; V.I.R. supervised and cowrote the manuscript; and all authors participated in writing and revising the manuscript to its final form.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Vivienne I. Rebel, Greehey Children’s Cancer Research Institute, University of Texas Health Sciences Center at San Antonio, San Antonio, TX 78229-3900; e-mail: rebel@uthscsa.edu.