Abstract
Defining features of chronic lymphocytic leukemia (CLL) are not only its immunophenotype of CD19+CD5+CD23+sIgdim expressing clonal mature B cells but also its highly variable clinical course. In recent years, advances in massively parallel sequencing technologies have led to rapid progress in our understanding of the CLL genome and epigenome. Overall, these studies have clearly demarcated not only the vast degree of genetic and epigenetic heterogeneity among individuals with CLL but also even within individual patient leukemias. We herein review the rapidly growing series of studies assessing the genetic and epigenetic features of CLL within clinically defined periods of its growth. These studies strongly suggest an evolving spectrum of lesions over time and that these features may have clinical impact.
Introduction
Chronic lymphocytic leukemia (CLL) is an initially slow-growing common B-cell malignancy whose hallmark is a highly variable clinical course. For more than a decade, “watch and wait” has been the standard approach for patients without symptomatic disease, with frontline chemotherapy-based therapy as the conventional choice if treatment is required. Over the past 2 years, however, three new drugs have been approved: (1) the novel potent CD20-targeting antibody obinutuzumab,1 (2) the inhibitor of PI3-kinase idelalisib,2 and (3) the irreversible inhibitor of Bruton tyrosine kinase ibrutinib.3 Moreover, several highly active agents, such as the BCL2 inhibitor GDC-0199/ABT-199,4 are in advanced clinical trials, which further promise to expand treatment options.
With these growing therapeutic possibilities, understanding the heterogeneous features of this disease and how it evolves over time becomes a priority, so that maximum benefit can be gleaned from these diverse therapies. In parallel with the exciting transformation in the therapeutic landscape of CLL, an explosive growth in our understanding CLL genetics has taken place.5 Several large-scale studies of massively parallel sequencing have defined the mutation spectrum6-12 and have investigated the altered epigenome of CLL.13-18 Altogether, these studies have uncovered both the vast genetic and epigenetic heterogeneity among patients, and within individual patient samples. Indeed, recent work has demonstrated intratumoral heterogeneity to impact individual evolutionary trajectories and clinical outcome in CLL.12,18 These innovations afford the insight that clonal heterogeneity likely fuels clonal evolution and can contribute to the variability in clinical course among CLL patients.
Herein, we review recent progress in our understanding of the complexities of inter- and intratumoral genetic and epigenetic heterogeneity in relationship to evolutionary principles. These new data offer fresh perspectives on our approaches to the prognostication and management of CLL.
The extensive genetic heterogeneity of CLL
Studies of somatic copy number variations using karyotyping, fluorescence in situ hybridization or single nucleotide polymorphism arrays first revealed the high molecular heterogeneity of CLL (extensively reviewed elsewhere).19,20 The most recurrent lesions identified were deletions of chromosome 13q (55% of cases), 17p (7%), and 11q (6% to 18%); and trisomy 12 (12% to 16%).21,22 In addition to their prognostic relevance, the minimal deleted region of each of these deletions have been found to contain within them putative CLL drivers: ATM and BIRC3 in 11q, TP53 in 17p, and miR-15a/16 encoded in an intron of DLEU2 in 13q.23
In recent years, the relative affordability of next-generation sequencing (NGS)-based technologies such as whole-genome and whole-exome sequencing has made it feasible to undertake large-scale efforts in cancer sequencing. A primary goal of the initial forays to dissect the CLL genome was to discover, in an unbiased fashion, new drivers based on statistical modeling. Rapid strides in our genetic understanding were gained because of several salient features of CLL that facilitated genomic investigation; namely, the ready accessibility of purified tumor cells from peripheral blood, its relatively indolent kinetics allowing for repeated sampling over time to study disease evolution and the highly variable clinical courses of patients that provides a strong distinguishing signal from which the impact of novel features can be differentiated.
Collectively, the earliest NGS-based sequencing studies revealed the overall low somatic mutation rate in CLL (∼1/Mb), similar to other hematologic malignancies, but at least 10-fold lower than carcinogen- or UV-induced solid tumors (∼15/Mb for melanoma).24 These studies demonstrated the highly heterogeneous genetic nature of CLL, characterized by several “mountains” (ie, significantly recurrent genes) but also “hills” (ie, infrequently recurrent genes), with the lack of any discernible universal genetic event accounting for all cases.6-11 These early studies of up to ∼100 cases each (with clinically heterogeneous sample cohorts) corroborated known CLL-associated alterations, such as somatic mutations, across the length of the DNA damage response genes TP53 and ATM, consistent with their inactivating effect. Unexpectedly, these studies further uncovered a number of novel frequent somatic changes (likely activating at hot spot locations). Mutations in the PEST domain of the key ligand-activated transcription factor of the NOTCH signaling pathway NOTCH1 (c.7544_7545fsdel) were among the first novel alterations to be discovered by NGS.7 Also discovered were the recurrent L265P mutations in the critical adaptor of the toll-like receptor complex MYD88, leading to potential constitutive nuclear factor-κB signaling. Finally, mutations in the essential splicing factor SF3B1 were identified, localized to evolutionarily conserved hotspots within its carboxyl-terminal repeat HEAT domains (most frequently at K700E).6,9 SF3B1 is a central component of the U2 spliceosome, which orchestrates the excision of introns from pre-messenger RNA (mRNA) to mature mRNA.25
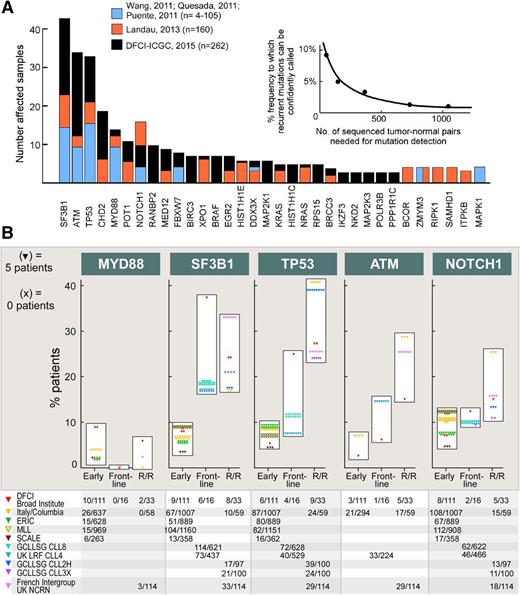
Subsequent to these initial reports, studies with larger-sized cohorts have uncovered additional novel candidate drivers including new chromatin regulators (CHD2 and HIST1H1C), B-cell transcription factors (EGR2 and IKZF3), RNA export factors (XPO1 and RANBP2), ribosomal proteins (RPS15), telomere-associated proteins (POT1), and signal transducers (RAS, MAP2K1, and MAP2K3) (Figure 1A).10,12,26 Altogether, these studies underscore the importance of sufficient power for sensitive detection of drivers in this highly genetically heterogeneous disease. Based on a saturation analysis and taking into account the background mutation rate of CLL, it has been estimated that an analysis of ∼2000 samples would be sufficient to confidently identify recurrent drivers present in 1% to 2% of the population. Hence, as expected, with the incremental growth in size of the discovery cohort, we have observed a growing “long tail” of significant drivers (Figure 1A).24,26
The mutational landscape of CLL. (A) The discovery of recurrently CLL mutated genes has become more sensitive with increased cohort size, with the estimated sensitivity calculated through saturation analysis6,12,26 ; (B) Frequency of gene mutations depending on the course of the disease. “Early” (newly diagnosed and untreated patients); “Frontline” (untreated patients with symptomatic CLL requiring therapy); and “R/R” (relapsing or refractory patients). Unselected cohorts have been included from: DFCI/Broad Institute,12 Amedeo Avogadro University of Eastern Piedmont, Novara and Sapieza University (Rome, Italy)/Columbia University (New York)27 ; ERIC 28 ; MLL29 ; and SCALE.30 Reported are also series from clinical trials: UK LRF CLL431,32 ; GCLLSG CLL8,33 CLL2H,34 and CLL3X35 ; GCFLLC/MW-GOELAMS ICLL01, and UK NCRN CLL201 and CLL202.36 DCFI, Dana-Farber Cancer Institute; ERIC, European Research Initiative on CLL; GCFLLC/MW-GOELAMS, French CLL Intergroup; GCLLSG, German CLL Study Group; MLL, Munich Leukemia Laboratory; SCALE, Scandinavian Lymphoma Etiology; UK LRF, United Kingdom Lymphoma Research Foundation; UK NCRN, United Kingdom National Cancer Research Network.
The mutational landscape of CLL. (A) The discovery of recurrently CLL mutated genes has become more sensitive with increased cohort size, with the estimated sensitivity calculated through saturation analysis6,12,26 ; (B) Frequency of gene mutations depending on the course of the disease. “Early” (newly diagnosed and untreated patients); “Frontline” (untreated patients with symptomatic CLL requiring therapy); and “R/R” (relapsing or refractory patients). Unselected cohorts have been included from: DFCI/Broad Institute,12 Amedeo Avogadro University of Eastern Piedmont, Novara and Sapieza University (Rome, Italy)/Columbia University (New York)27 ; ERIC 28 ; MLL29 ; and SCALE.30 Reported are also series from clinical trials: UK LRF CLL431,32 ; GCLLSG CLL8,33 CLL2H,34 and CLL3X35 ; GCFLLC/MW-GOELAMS ICLL01, and UK NCRN CLL201 and CLL202.36 DCFI, Dana-Farber Cancer Institute; ERIC, European Research Initiative on CLL; GCFLLC/MW-GOELAMS, French CLL Intergroup; GCLLSG, German CLL Study Group; MLL, Munich Leukemia Laboratory; SCALE, Scandinavian Lymphoma Etiology; UK LRF, United Kingdom Lymphoma Research Foundation; UK NCRN, United Kingdom National Cancer Research Network.
The most commonly mutated genes (TP53, SF3B1, MYD88, NOTCH1, and ATM) have remained unchanged across studies. However, their reported frequencies across studies have been variable, related to the variable incidence of each particular mutation from the early stages of disease to the time of first therapy, or even at relapse (Figure 1B).12,27-36 SF3B1 mutations appear to associate with early progression, with its frequency increasing from 4% to 9% at diagnosis to 17% to 18% or greater by the time of first therapy. By contrast, the frequency of MYD88 is unchanging across disease stages, whereas NOTCH1 mutation increases mainly between the time of first therapy (∼10%) and relapse (up to 25%). Finally, mutations in TP53 and ATM each rise continuously through the course of disease, from a frequency of <10% in early disease to 25% or greater at relapse. Overall, whereas the variable frequencies of CLL drivers may reflect the association of certain gene mutations to other aggressive CLL features (eg, unmutated immunoglobulin heavy chain variable [IGHV] genes), these differing trajectories may also suggest potentially different roles of these drivers over the CLL course.
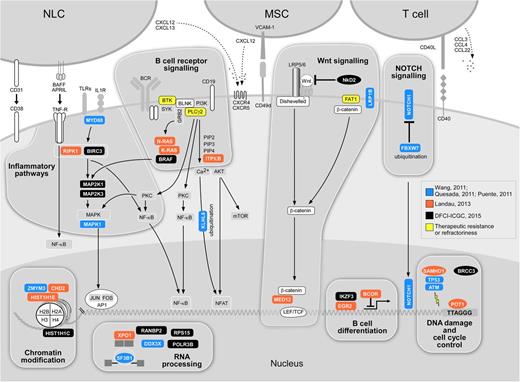
The central roles of the mutated genes in several essential cellular processes pathways have suggested these as core CLL pathways. Indeed, as the experience in characterization of the mutational landscape has expanded, greater resolution for mapping the nodes to which CLL might be sensitive has been achieved. As shown in Figure 2, CLL mutations have been consistently observed to involve pathways in DNA damage (TP53 and ATM), mRNA processing (SF3B1 and XPO1), chromatin modification (HIST1H1E, CHD2, and ZMYM3), Wnt signaling, NOTCH signaling (NOTCH1), and inflammation (MYD88). More recently, novel drivers further support somatic mutation as a mechanism affecting B-cell–related signaling and transcription (EGR2 and BRAF).37 The functional role of several novel putative drivers has been confirmed across several studies. For example, through an innovative delivery system for nucleic acids into CLL cells, the silencing of mutated Wnt pathway genes in cells harboring those mutations could be effectively achieved, and demonstrated a loss in the viability of these cells, suggesting dependence of those CLLs on Wnt pathway signaling.38 Of note, several novel drivers appear to involve the disruption of the DNA damage response. Mutations in POT1, involved in the protection of telomeres, were confirmed to prevent its binding to telomeric DNA and to result in the generation of numerous telomeric and chromosomal abnormalities.10 Mutations in SF3B1 have been shown to lead to altered splicing,6,9,39 and was recently linked to an altered DNA damage response.40 SAMHD1, a regulator of the intracellular dNTP pool, has been demonstrated to be recruited to the site of DNA damage and is likely involved in the response to DNA double-strand breaks.41 It should be noted that CLL cells are exquisitely sensitive to microenvironmental cues,42 and hence future studies will undoubtedly evaluate the impact of genetic drivers and pathways on the functional behavior of CLL cells in relationship to immune and stromal cell populations with which they interact as well as treatment received.43,44
Putative core cellular pathways affected by significantly mutated genes in CLL. Blue-genes identified from CLL series by Wang et al,6 Puente et al,8 and Quesada et al9 (n = 4 to 105 samples); orange-genes identified by Landau et al (n = 160 subjects)12 ; black-genes affected by 262 subjects23 ; yellow gene mutations identified in relationship to drug resistance.43,44 Several of the affected pathways likely serve as an important bridge with the microenvironment, which is of particular importance in CLL (crucial actors of the CLL microenvironment are represented by NLCs, MSCs, and T cells. NLCs, nurse-like cells; NF-κB, nuclear factor-κB; MSCs, marrow stroma cells; mTOR, mammalian target of rapamycin; TLR, toll-like receptor.
Putative core cellular pathways affected by significantly mutated genes in CLL. Blue-genes identified from CLL series by Wang et al,6 Puente et al,8 and Quesada et al9 (n = 4 to 105 samples); orange-genes identified by Landau et al (n = 160 subjects)12 ; black-genes affected by 262 subjects23 ; yellow gene mutations identified in relationship to drug resistance.43,44 Several of the affected pathways likely serve as an important bridge with the microenvironment, which is of particular importance in CLL (crucial actors of the CLL microenvironment are represented by NLCs, MSCs, and T cells. NLCs, nurse-like cells; NF-κB, nuclear factor-κB; MSCs, marrow stroma cells; mTOR, mammalian target of rapamycin; TLR, toll-like receptor.
Intratumoral genetic heterogeneity provides insights into the order of mutation acquisition in CLL
Despite its clonal origin, cancer is characterized by the coexistence of multiple populations within the tumor. Such intratumoral heterogeneity was conceptualized decades ago as an inevitable outcome of the mutational process inherent to cancers45,46 and could be detected using a variety of experimental methods.47,48 NGS, however, has been transformative for this effort by providing a comprehensive approach to detect subclones at unprecedented resolution.49-51 For CLL, the relative high purity of samples has facilitated the confident detection of rare allelic frequencies of somatic variants, which upon correction for local ploidy and clustering, can lead to the defining of subclonal populations within tumor samples, and hence have led to the understanding of the pervasive extent of intratumoral heterogeneity present in CLL.
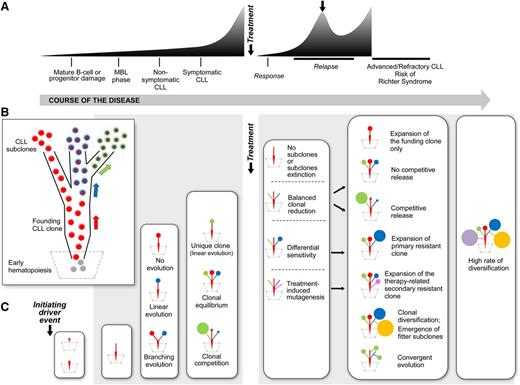
The ability to define subclonal architecture in CLL by tumor sequencing has suggested an ability to infer the phylogeny of any case, since the “snapshot” provided by subclonal composition likely results from the stepwise acquisition of mutations across the process of leukemogenesis (Figure 3A-B). Conceptually, the earliest clone-propagating event would have occurred in a single cell, giving rise to a “CLL-founding clone.” Thus, within this framework, clonal mutations within a bulk sample represent earlier events, acquired as putative cancer-initiating or driving events or alternatively, as passenger events that were present at the time of transformation. Landau et al found del(13q), trisomy 12, and MYD88 mutations as the three most significantly clonal lesions; consistent with their acquisition early in the history of individual CLL tumors, these could potentially provide clonal advantage to B cells.12 Mutations in NOTCH1 and SF3B1 were also commonly clonal. Applying machine-learning–based approaches to large cross-sectional CLL datasets, it has been proposed that the acquisition of these early CLL drivers then leads to preferred evolutionary trajectories.52
Heterogenous evolutionary trajectories through CLL course and therapeutic intervention. (A) Typical CLL disease course; (B) phylogenetic tree of CLL leukemogenesis (each arrow represents the acquisition of a genetic event); (C) evolution of the CLL phylogenetic tree at various stages of the CLL disease course.12,48,58-61
Heterogenous evolutionary trajectories through CLL course and therapeutic intervention. (A) Typical CLL disease course; (B) phylogenetic tree of CLL leukemogenesis (each arrow represents the acquisition of a genetic event); (C) evolution of the CLL phylogenetic tree at various stages of the CLL disease course.12,48,58-61
Consistent with the idea that early alterations can provide a clonal advantage to B cells, Klein et al demonstrated that as a lowly penetrant lesion, B cells restricted expression of del(13q) could give rise to histopathologic evidence of CLL in a murine model.53 Moreover, recent studies have demonstrated that many cancer-driving genetic events were detectable in large population-based cohorts without hematologic malignancies, the so-called “clonal hematopoiesis.”54,55 The frequency of these events increased with age, and although most mutations occurred in DNMT3A, TET2, and ASXL1 (associated with myeloid malignancies), significant events were also noted in TP53 and SF3B1 as well as rare instances of mutations in MYD88 and NOTCH1. The presence of clonal hematopoiesis was associated with an increased risk of various hematologic cancers including CLL. More directly evaluating early hematopoiesis in CLL, Damm et al applied targeted deep sequencing of candidate early driver mutations on flow-sorted hematopoietic progenitors, and intriguingly, detected the presence of lymphoid oncogenes (ie, mutated BRAF, NOTCH1, and SF3B1) in CD34+ hematopoietic cells.37 Together with prior murine xenograft studies demonstrating that hematopoietic stem cells (HSCs) from CLL patients can generate clonal B cells with CLL-like phenotype,56 these data suggest hematopoietic progenitors as cells-of-origin for CLL.
Intratumoral genetic heterogeneity fuels diverse evolution patterns in CLL
Clonal evolution refers to the process in which cancer cells present accumulated genetic (and epigenetic) changes over time giving rise to new subclones. Cancer is thought to evolve by a process of clonal expansion, diversification, and selection within the tissue ecosystems.46 This feature appears to be in line with an evolutionary process as those reported by Darwin about species evolution.
A key challenge presented by intratumoral heterogeneity is its capacity to fuel clonal evolution and the generation of therapy resistant subpopulations. Through acquisition of fitter subclones and environmental selective pressures, tumors can evolve with time. Indeed, the presence of subclonal driver mutations as a surrogate of an active evolutionary process driving of clonal diversification was found to be an independent poor prognostic risk factor of more aggressive and/or resistant disease.12,57
Genetic changes across the disease course of CLL has been investigated in a growing body of studies, which have used longitudinal follow-up of the subclonal composition by various genomic technologies and have illustrated the patterns of clonal evolution across the natural history of CLL (Figure 3C).12,48,58-61
Monoclonal B-cell lymphocytosis (MBL) is thought to be the premalignant lesion of CLL.62 Although only a small number of MBL cases have been characterized by whole-exome sequencing, a broad heterogeneous spectrum of mutations are clearly present in MBL (including SF3B1, NOTCH1, FBXW7, and DDX3X), similar to the mutational spectrum of CLL.63 In the majority of cases reported thus far, trisomy 12 and del(13q) were also detected by fluorescence in situ hybridization, supporting the idea that these are early lesions in CLL.
Overall, two common patterns of clonal evolution in CLL have been observed (Figure 3C).12,48,58-61 One is that of linear evolution, in which a single clone undergoes successive acquisition of additional driver events over time, that accumulate atop the initial abnormalities. The other is that of branched evolution, in which two or more genetic subclones coexist and evolve in a parallel fashion. In general, inference of subclonal architecture in bulk single time point samples using algorithmic approaches cannot necessarily resolve linear vs branched structures of subclonal populations, although recent in silico models have attempted to explore potential evolutionary paths from early to secondary events.52 Emerging studies evaluating the role of ultra-deep sequencing on the one hand and novel technologies to dissect the mutational profiles of single cells on the other hand, promise to provide further insights on CLL clonal architecture and evolution.64-66
By now, it has become clear that multiple subclones can maintain their relative proportions to each other over years (“clonal equilibrium”), whereas in other cases, individual subclones can emerge as dominant over time, likely because of their relative higher fitness (“clonal competition”) (Figure 3C). The proportion of patients in each of these two categories is likely dependent of stage of disease.
The concepts of clonal equilibrium vs competition and evolution have been most evident in longitudinal studies assessing the impact of cytoreductive therapy.12,48,58-61 Landau et al suggested that the absence of intervening therapy was largely associated with stable subclonal composition over time. In contrast, chemotherapy exposure predominantly resulted in marked clonal evolution.12 Across studies, the genetic lesions dominating at relapse have been consistently detected in smaller subclones at earlier stages, suggesting the emergence of fitter subclones together with genetic diversification.52 The most consistent lesion detected in these studies has been preexisting TP53 mutations, resulting in the outgrowth of highly genomically complex clones at relapse.48 On one hand, genetic lesions such as mutated TP53 itself may confer resistance to the therapeutic agent, and a differential sensitivity of the subclone to these drugs may be responsible for its dominance at relapse. Alternatively, the emergence of a dominant subclone could result from a balanced clonal reduction, followed by a competitive release that depends on subclone-related growth properties and mechanisms of tumor progression. Finally, therapy itself could induce de novo mutations conferring major fitness of the related subclones (Figure 3C).67
The notion that a coherent pattern of resistance emerging following exposure to therapy has been most clearly demonstrated by recent studies describing resistance developing after exposure to potent inhibition of B-cell–receptor signaling with ibrutinib. Resistance was linked to mutations in Bruton tyrosine kinase (C481S) and/or its immediate downstream partner PLCγ2 (Figure 2).43 Remarkably, various individual subjects across studies have been detected to harbor multiple subclones with mutation in PLCγ2, consistent with convergent evolution occurring within these patients.43,59,61,68 These findings support the idea of individualized evolutionary trajectories taken on by different CLL subclones to functionally circumvent the pathway inhibition imposed by targeted inhibitors.
Profiling CLL epigenetic heterogeneity
In addition to genetic lesions, the disruption of epigenetic mechanisms also plays a role in oncogenesis.69 DNA methylation, which occurs at the cytosine residue of the CpG dinucleotide, is a crucial facet of epigenetic programming that normally regulates gene transcription and genome stability, and contributes to normal B-cell development.70 Although potentially reversible, DNA methylation status is considered an inheritable trait. Furthermore, the CLL methylome over time has been characterized as rather stable, with few changes identified between the resting and proliferating compartments of CLL.16 On the other hand, in striking support for a key role of epigenetics contributing to CLL leukemogenesis, Chen et al reported the detection of aberrant methylation well before disease onset in the Eμ-TCL1 mice, an established murine model of CLL.71,72 Furthermore, aberrant promoter methylation leading to dysregulated expression of genes such as TCL1,73 DAPK1,74 or ZAP70,75 as well as CLL-associated microRNAs76,77 and long intervening noncoding RNAs78 have been implicated in CLL pathogenesis.
Comprehensive methylation profiling by genome-wide arrays (high-resolution Illumina 450K) or sequencing (whole-genome bisulfite sequencing) have provided a global picture of methylation changes in CLL compared with normal B cells, and have demonstrated the highly heterogeneous methylation profiles across samples. Similar to other cancers,79-81 CLL harbors global genome-wide hypomethylation, with localized regions of hypermethylation.13-18 Among CLL patients, differences in methylation patterns, most of them lying outside CpG islands, have been clearly observed in relation to IGHV mutational status.13,16 In addition, based on similarities in methylation imprint, these systematic investigations have suggested naïve B cells as the putative cell-of-origin of CLLs with unmutated IGHV, and memory B cells for CLLs with mutated IGHV.15 In multivariate analyses, methylation imprint also influenced time-to-treatment.
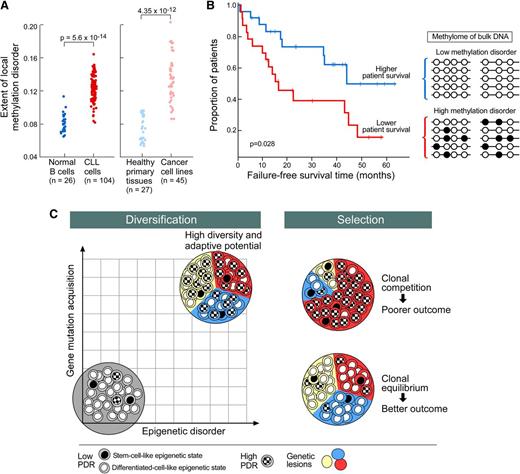
More recently, the extensive intratumoral epigenetic heterogeneity in CLL and its impact of clonal evolution was investigated. Oakes et al reported that, unlike normal B cells, CLL cells harbored greater intermediate DNA methylation values, which were attributed to allele-specific methylation.17 The estimated high level of intratumoral methylation heterogeneity was associated with aggressive features and a shorter time-to-first treatment. Furthermore, evolution of DNA methylation over time, observed in 9 of 28 cases, was linked to a higher level of methylation heterogeneity at the earlier time point and to the presence of subclonal (rather than clonal) genetic events, thus demonstrating a link between genetic and methylation evolution.
Using reduced representation bisulfite sequencing, Landau et al also detected increased intermediate DNA methylation values, but pervasive locally disordered methylation (assessed by proportion of discordant reads [PDR]) throughout the genome was the primary basis of CLL intratumoral methylome heterogeneity.18 This high level of epigenetic “noise” appeared to arise stochastically and was specific to cancer (rather than a change related to normal tissue differentiation) (Figure 4A).18,82 Locally disordered methylation impacted the variability of gene expression across and within samples, creating enhanced potential for alternative evolutionary trajectories. In particular, locally disordered methylation preferentially affected genes associated with stem cell biology and hence could provide fuel for the potential subclonal diversification of leukemic cells. High promoter PDR was associated with shorter failure-free survival independently of other risk factors (Figure 4B)18 and samples with higher promoter PDR were more likely to have a subclonal driver mutation. Hence, methylation disorder may, together with genetic instability, contribute to the clonal diversification process (Figure 4C). Future studies may elucidate the extent to which locally disordered methylation plays a role in CLL initiation, progression, and therapeutic resistance.
Model of clonal diversification and selection in CLL. (A) Locally disordered DNA methylation is higher in CLL and cancer tissues compared with normal tissues, including B cells (adapted from Landau et al18 ); (B) Adverse impact of locally disordered DNA methylation on failure-free survival18 ; (C) Clonal diversification may result from synergic effects of disordered DNA methylation and genomic instability (left). Clonal selection related to therapeutic and microenvironment pressure shapes the subclonal composition that ranges from clonal equilibrium to clonal competition (right). High level of clonal diversification could lead to the emergence of fitter subclones competing within CLL bulk and is responsible for more aggressive/resistant disease associated with poor survival.
Model of clonal diversification and selection in CLL. (A) Locally disordered DNA methylation is higher in CLL and cancer tissues compared with normal tissues, including B cells (adapted from Landau et al18 ); (B) Adverse impact of locally disordered DNA methylation on failure-free survival18 ; (C) Clonal diversification may result from synergic effects of disordered DNA methylation and genomic instability (left). Clonal selection related to therapeutic and microenvironment pressure shapes the subclonal composition that ranges from clonal equilibrium to clonal competition (right). High level of clonal diversification could lead to the emergence of fitter subclones competing within CLL bulk and is responsible for more aggressive/resistant disease associated with poor survival.
Potential prognostic impact of the CLL genomic features
Cytogenetic evaluation remains the gold standard and basis for the long-standing hierarchical classification of CLL.21 However, given that several of the novel CLL-associated gene mutations individually have been reported to hold prognostic significance, Rossi et al integrated mutational and cytogenetic information from a heterogeneous series of 637 CLL patients (with further validation using 370 patients), and found that the addition of molecular information improved prognostication of overall survival (OS) compared with cytogenetics alone.27 Overall, this schema distinguished 4 subgroups: high-risk (TP53 and/or BIRC3 abnormalities), intermediate-risk (NOTCH1 and/or SF3B1 mutations and/or del[11q]), low-risk (+12 or normal genetics), and very low-risk (del[13q] only). Similarly, Jeromin et al assessed cytogenetic information with a panel of common CLL-associated mutations in 1160 untreated CLL patients (82.6% of cases from diagnosis).29 These analyses highlight the concept that refinement of prognostic schema is possible with the addition of gene mutation information. In these studies, the presence of SF3B1 mutations was confirmed as a predictor of significantly shorter time-to-first therapy (3.8 vs 8 years), even within a multivariate model including unmutated IGHV and del(11q).29 Likewise, the European Research Initiative on CLL recently conducted a multivariate analysis including 774 patients evaluable for time-to-first therapy and confirmed the adverse influence on outcome of mutated SF3B1, unmutated IGHV, and del(11q), in addition to TP53 disruption.28 SF3B1 mutations further retained its poor prognostic impact on OS, in addition to TP53 disruption.29
A growing body of studies have evaluated the impact of a limited number of recurrent CLL-associated gene mutations (TP53, NOTCH1, and SF3B1) on outcome in the setting of clinical trials.32-35 The impact of these mutations has been variable across these studies and likely reflects the dependence of the impact of these alterations on when in the course of disease they are evaluated. For example, Stilgenbauer et al evaluated more than 600 of 817 previously untreated patients enrolled on the German CLL Study Group CLL8 trial, and upon multivariate analysis, found that OS was impacted by TP53 mutation status but not by SF3B1 or NOTCH1 mutations, whereas PFS was adversely affected by SF3B1 and TP53 mutations.33,83 Patients with NOTCH1 mutations did not benefit from rituximab. By contrast, analyses of the United Kingdom Lymphoma Research Foundation CLL4 trial subjects revealed TP53 disruption, NOTCH1 mutation, and SF3B1 mutations as all significantly impacting OS.32 Two further analyses of clinical trial cohorts (CLL2H–alemtuzumab for fludarabine-refractory patients and CLL3X–reduced-intensity HSC transplantation in poor-risk patients) failed to show any adverse impact of NOTCH1 or SF3B1 mutations on response rate, PFS, or OS after multivariate analysis.34,35 Further research studies will be required to definitively establish the role of individual driver mutations on the response to specific therapies.
Incorporating evolutionary concepts into schema for CLL prognosis, monitoring, and treatment
The tracking of intratumoral heterogeneity over time can provide critical information on clonal dynamics, which could impact the management of patients. Because the dominant subclones at relapse may be present as minor subclones earlier in disease course, it follows that early detection of these subclones of known high fitness could lead to the contraindication of treatment known to select such clones. One striking example is that of somatic mutations disrupting TP53. Small TP53 mutated subclones identified before treatment appear to anticipate the dominant population at relapse.84 Presence of mutations leading to TP53 disruption clearly predicts poor response to frontline chemotherapy. Possibly, patients with subclonal TP53 disruption will respond better to novel potent targeted inhibitors (ie, ibrutinib and idelalisib).85-87 In a separate example, recent mathematical modeling of the kinetics of ibrutinib resistance has suggested that such resistant clones are present at the time of treatment initiation.88 Hence, high-resolution molecular characterization before and during therapy has the potential to detect disease relapse in advance of full hematologic evidence of resistance.
From the standpoint of therapy, the presence of intratumoral heterogeneity strongly supports on the one hand personalized therapies, and on the other hand, supports the use of therapies that can target multiple vulnerable nodes simultaneously, whether this is through combination therapy that can include chemotherapy and/or combined targeted inhibition, or even immunotherapy.89
Because clonal equilibrium has been linked to stable disease, a provocative approach for the monitoring and treatment of CLL could be efforts to lead to a more stable state (as observed in early stages patients). The assessment and monitoring of clonal heterogeneity might be a useful tool to appreciate both the magnitude and the quality of the residual disease. For example, stabilization of the epigenome, which has been recently reported to fuel clonal evolution and diversity in CLL, is now feasible with newly available agents and has not been standardly tested in CLL, and yet may have an impact on limiting the aggressiveness of progressive evolutionary sweeps. Thus, as an “adaptive therapy” strategy,90 a key goal would be to maintain subclonal relationships such that the emergence of the more resistant clones is prevented. Another potential therapeutic opportunity is presented by the recent data arguing for the occurrence of cancer-driving events at the level of hematopoietic progenitor cells in CLL. Certainly, these findings may explain why, so far, allogeneic HSC transplantation is the sole strategy that has demonstrated curative potential in CLL.91,92 Furthermore, active immunotherapy, a now maturing field,93 has the capacity to impose evolutionary pressure distinct from conventional therapies, and which, if applied early enough in disease course, can modify the natural history of this disease.
Conclusion
The advent of genome-wide sequencing technologies has uncovered the tremendous genetic and epigenetic heterogeneity of CLL. The extent to which these features should be taken into account in the management of patients remains to be seen. The identification of novel genetic drivers, which has led to the characterization of interpatient and intratumoral heterogeneity has been shown to clearly impact clinical outcome. Furthermore, a growing series of longitudinal analyses across the stages of CLL disease has delineated not only the heterogeneous trajectories of clonal evolution but also their close link to therapeutic pressure. At the present time, with the ever-increasing availability of effective chemotherapy, immunotherapy, and agents for the targeted inhibition of key CLL pathways, it is evident that each therapeutic modality provides its own unique mode of selective pressure on a population of CLL cells. Further detailed study in each of these areas using existing or novel (including single-cell approaches) genome-wide technologies, together with functional analyses in vitro and in vivo, will certainly inform us of the relationship between the subclonal architecture in CLL and nodes of therapeutic resistance, and hence provide the critical knowledge gap required for further developing improved individualized and effective therapies for CLL patients.
Acknowledgments
C.J.W. acknowledges support from the Blavatnik Family Foundation, American Association for Cancer Research (SU2C Innovative Research Grant), National Institutes of Health, National Heart, Lung, and Blood Institute (1RO1HL103532-01 and 1RO1HL116452-01), National Cancer Institute (1R01CA155010-01A1), and the Lymphoma Research Foundation. C.J.W. is also a recipient of a Leukemia Lymphoma Society Scholar Award and Translational Research Program Award, as well as a Quest for Cures Award.
Authorship
Contribution: R.G. and C.J.W. conceived the article, wrote the manuscript, and generated the figures.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Catherine J. Wu, Harvard Medical School, Department of Medical Oncology, Dana-Farber Cancer Institute, 450 Brookline Ave, DA 540B, Boston, MA 02115-5450; e-mail: cwu@partners.org.