Abstract
Monocytes are recruited from peripheral blood to inflamed tissues during infections and wound healing. Monocyte adaptation to hypoxia is essential in inflamed tissues which are characterized by low oxygen tensions. Monocytes respond to hypoxia by widespread changes in transcript levels. We have also recently described that hypoxia induces site-specific C>U RNA editing of hundreds of genes in monocytes, which is mediated by the innate antiviral restriction factor APOBEC3A (A3A) cytidine deaminase. However, mechanisms regulating hypoxia-sensing and signaling in monocytes are poorly understood. Here we show that inhibition of mitochondrial complex II (succinate dehydrogenase; SDH) by the ubiquinone-analogs atpenin A5 (AA5) or TTFA induces hypoxic gene expression and A3A-mediated RNA editing in normoxic cultures of monocytes. RNA seq and RNA editing analyses show that normoxic treatment of monocytes with AA5 closely mimics the hypoxia effects in inducing transcriptome-scale changes (Figure 1). RT-qPCR validation experiments confirm that normoxic treatment of monocytes with AA5 induces A3A-mediated RNA editing and causes upregulation of many hypoxia-related and pro-angiogenic genes including VEGFA, CXCL8 (IL8) and CXCR4. Treatment of monocytes with AA5 in normoxia further enhances the interferon-induced cytidine deaminase enzyme activity of A3A. AA5 does not inhibit A3A-mediated RNA editing or transcript induction in hypoxic monocytes. In contrast, myxothiazol, a complex III inhibitor, does not induce A3A-mediated RNA editing in normoxia and inhibits it in hypoxia. Thus, monocyte response to hypoxia is specifically triggered by inactivation of mitochondrial complex II. Previous studies on familial paragangliomas, pseudohypoxic highly vascularized neuroendocrine tumors which are associated with germ line SDH mutations, have suggested stabilization of HIF1 alpha in mediating the hypoxia-induced gene expression. In contrast, we find that normoxic treatment with AA5 does not stabilize HIF-1 alpha or HIF-2 alpha in monocytes or in 293T embryonic kidney cells (Figure 2). Moreover, forced stabilization of HIF-1 alpha by treatment of monocytes with DMOG blocks hypoxic induction of A3A-mediated RNA editing. Treatment of monocytes with small molecule inhibitor of HIF-1 alpha (CAY10585) or with an HIF-2 alpha antagonist (SML0883) does not inhibit hypoxic induction of A3A-mediated RNA editing. Taken together, our findings identify complex II inhibitor AA5 as a hypoxia mimetic that enhances the interferon-inducible RNA editing activity of the antiviral enzyme A3A in monocytes and suggest that oxygen sensing by SDH controls transcriptional responses to hypoxia through a distinct signaling pathway that does not involve HIF-1 alpha or HIF-2 alpha.
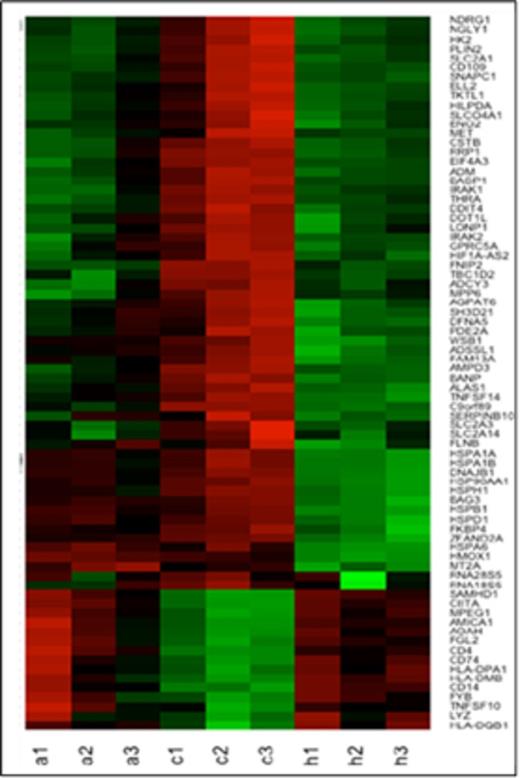
Heat map shows that the most differentially expressed genes (listed on the right) between normoxic control (c1,c2,c3) and hypoxic (1% O2) monocytes (h1,h2,h3) (defined as FDR<0.05, log2 fold change >3 and average expression >6 cpm) also show similar changes in normoxic monocytes exposed to AA5 (a1, a2, a3). (green high/red low expression).
Heat map shows that the most differentially expressed genes (listed on the right) between normoxic control (c1,c2,c3) and hypoxic (1% O2) monocytes (h1,h2,h3) (defined as FDR<0.05, log2 fold change >3 and average expression >6 cpm) also show similar changes in normoxic monocytes exposed to AA5 (a1, a2, a3). (green high/red low expression).
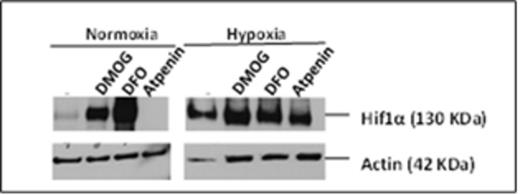
Western blots show that in contrast to DMOG (1 mM) and DFO (0.5 mM), which are known to stabilize HIF-1 alpha, atpenin A5 (1 μM) does not stabilize HIF-1 alpha in normoxia in 293T cells. Actin bands are loading controls.
Western blots show that in contrast to DMOG (1 mM) and DFO (0.5 mM), which are known to stabilize HIF-1 alpha, atpenin A5 (1 μM) does not stabilize HIF-1 alpha in normoxia in 293T cells. Actin bands are loading controls.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal