Abstract
Targeting bone marrow stromal interactions in multiple myeloma as a way of disrupting survival niches in patients have been the focus of novel therapies that include drugs like lenalidomide, bortezomib and dexamethasone. Our recently published studies and our previous abstracts presented at ASH have focused on interactions between myeloma cells and their stromal partners, dendritic cells that express the CD28 ligand CD80 and CD86. But in myeloma cells, CD28 expression has been linked to poor prognosis and tumoral expansion in patients. Further, most MM cells also co-express the CD28 ligand CD86, but not CD80, and expression of CD86 has also been linked with poor survival and tumorigenesis in myeloma patients. All MM cell lines (that are also representative of extramedullary myeloma) that we and others have tested co-express CD28 and its ligand CD86 on their surface, suggesting that MM cells could potentially interact with each other. Very little is known about potential molecular interaction between MM cells themselves, which might help to explain how MM cells survive outside the bone marrow such as advanced extramedullary myeloma.
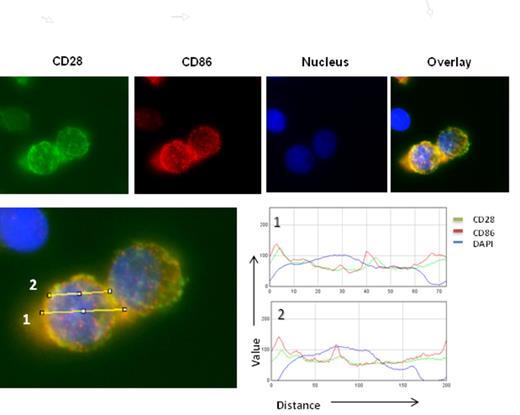
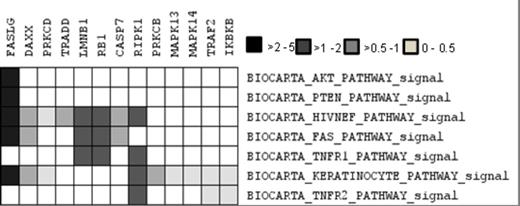
We have shown previously that CTLA4Ig treatment not only blocks MM interaction with DC in vitro but also sensitizes MM cells themselves to chemotherapy. We hypothesized that CTLA4Ig could be blocking CD28 -CD86 interactions between MM cells thus down regulating CD28 activation. But what we did not know was the nature of these interactions, and the downstream pathways regulated by CD28 activation. In this study, we present microscopic analysis and microarray gene expression studies to show that CD28 activation is dynamic and modulates several key pathways involved in survival and cell death. Interestingly we observed that CD28 on the surface of myeloma cells interact with its co-expressed ligand CD86 within pits and folds on the myeloma cell surface (Fig 1) apart from interactions with each other. Colocalization profiling using RGB profiler software (tool in ImageJ) suggests close overlaps between CD28 and CD86 profiles on MM cell surface suggesting in situ interaction between these two molecules. This phenomenon dissipated upon treating the cells with CTLA4Ig. To identify important genes that are regulated by CD28 activation in myeloma, we did gene expression analysis of untreated and CD28 antibody treated samples. Pathway analysis was performed followed by clustering Leading edge analysis (LEA) on the gene expression dataset of ranked genes against gene sets representing over 100 pathways (BioCarta). We identified several gene clusters that seemed to be shared among 20 highest scoring pathways. Many of these pathways (AKT, IL2, FAS, ERK, PTEN, TNFR1) are known to be associated with CD28 activity in T cells. We then looked at the prevalence of these pathways among the 5 gene clusters identified by LEA and identified gene clusters of interest. These pathways were further selected and leading edge analysis performed to more closely identify genes commonly regulated between them. Genes upregulated in CD28 activated MM.1S such as FASL (FAS ligand) that has a role in immune evasion in myeloma54, RIPK1 (RIP kinase1), TRAF2, DAXX, PRKCD, RB1, IKBKB all of which have important roles in the activation and regulation of NFkB activation, a key event previously shown to occur downstream of CD28 activation in myeloma. Activation of several MAPK proteins that are essential for myeloma survival were also observed. Upregulation of pro-survival cytokines and their receptors like IL-8 (1.7 fold) and its receptor IL-8RB (1.8 fold), IL-5 (1.7 fold), IL-33 (3.1 fold), IL-15 (1.9 fold), the IL-6 receptor IL-6R (1.6 fold) and essential chemokines like CXCL6 (3) and CXCR4 (1.7 fold)58 were also observed in CD28 activated samples. The chemokine receptor CXCR4 and its ligand CXCL12 are known to be important for MM interaction with Bone marrow. We did not see any difference in CXCL12 levels. This results suggests that CD28 activation is dynamic and regulates several survival pathways in myeloma. We also present studies that show that many of these results observed are also replicated invitro. Our study reveals a number of new pathways and targets that could be utilized for developing new directions in myeloma research.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal