Abstract
Extracorporeal photopheresis (ECP) is an effective frontline therapy for patients with leukemic cutaneous T-cell lymphoma (L-CTCL), but the mechanisms of action are not fully understood. To elucidate molecular mechanisms underlying the efficacy of ECP, we used Agilent Whole Human Genome Microarrays to examine blood transcriptional profiles in L-CTCL patients after ECP therapy.
Ten L-CTCL patients including 5 clinical responders and 5 non-responders were studied. Their peripheral blood was collected before ECP (baseline), at Day 2, and one month post-ECP. Total RNA extracted from peripheral blood mononuclear cells was assayed with Whole Human Genome Oligo Microarrays (4 × 44 K) (Agilent, Santa Clara, CA). The differentially expressed gene analysis (DGA) was done using the paired t-test with Benjamini- Hochberg correction (P value < 0.05) between post-ECP and baseline. The fold change of gene expression between post-ECP and baseline were calculated from the normalized values. Hierarchical clustering of differentially expressed genes was performed with the Pearson correlation. The DGA between responders and non-responders were cross-compared. Canonical biological pathways were identified using Ingenuity Pathway Analysis (IPA, Ingenuity Systems, Redwood City, CA).
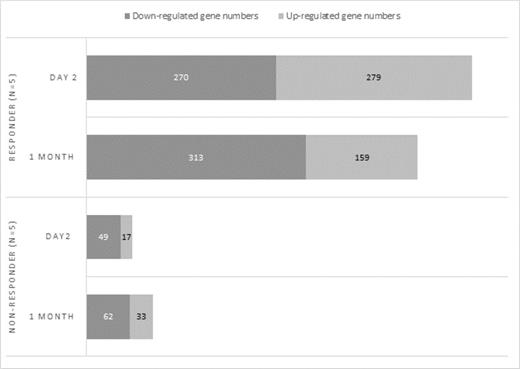
Differentially expressed gene profiles were different in responders from non-responders. As indicated in Figure 1, there were more genes differentially regulated in responders than in non-responders post-ECP at both Day 2 (549 genes in responders versus 66 genes in non-responders) and at one month (472 genes in responders versus 95 genes in non-responders). Among 472 differentially expressed genes in responders at one month post-ECP, almost twice as many genes (313) were down-regulated compared to up-regulated genes (159). The top down-regulated genes were IL-1β, EGR1, CCL3, CCL3L3, and CXCL2. The down-regulated genes were mainly related to functions of platelets, immune and/or stress responses, and chromatin remodeling. The upregulated genes were mainly related to functions of the nucleolus and included USP34, POLR3F, ZNF529, C22orf35, and BAT2D1. The ingenuity pathway analysis revealed that the top 5 pathways affected by ECP at one-month in responders were 1) integrin signaling; 2) granulocyte adhesion and diapedesis; 3) signaling by Rho Family GPTases; 4) agranulocytes (lymphocyte, monocyte and macrophage) adhesion and diapedesis; and 5) triggering receptor expressed on myeloid cells 1 (TREM1) signaling (Table 1). In contrast, these pathways and genes were less affected in non-responders. Of note, a comparison of all DGA results indicated that the responder group overlapped in the differentially expressed genes between Day 2 group (RD2) and one month group (RM1), but had few genes in common to the non-responder group (NM1). There were 94 genes consistently downregulated among RD2 and RM1 while only 6 genes were found in common between the RM1 and NM1 group. Similarly, 61 genes were consistently upregulated in group RD2 and RM1 while only 3 genes were found in common between the RM1 and NM1 group.
In summary, the blood transcriptional profiling by this study identifies a signature of genes and pathways relevant to clinical response to ECP in L-CTCL patients. These findings expand our understanding of molecular mechanisms of ECP. Further validation of these genes and pathways is warranted in the future studies.
Top canonical pathways affected by ECP in L-CTCL patients responded to ECP at one-month
| Canonical Pathways . | Downregulated genes . | Upregulated genes . |
|---|---|---|
| Integrin Signaling | 15/201 (7%) ITGA2B, MAP3K11, ITGA5, MYLK, ITGB3, MYL9, PARVB, AKT1, RHOB, CAPN1, ACTN4, CTTN, ARPC4, ACTN1, ITGB5 | 2/201 (1%) ITGB1, PPP1R12A |
| Granulocyte Adhesion and Diapedesis | 14/179 (8%) CSF3R, ICAM1, PPBP,ITGA5, CXCL5, SDC4, CCL3, ITGB3, GNAI2, CLDN5, CCL3L3, IL1B, CXCL1, CXCL2 | 1/179 (1%) ITGB1 |
| Signaling by Rho Family GTPases | 13/236 (6%) SEPT5, MAP3K11, ITGA5, MYLK, GNAZ, CDC42EP2, GNAI2, MYL9, GNG11, GNA15, RHOB, GNB2, ARPC4 | 3/236 (1%) ITGB1, DIAPH3, PPP1R12A |
| Agranulocyte Adhesion and Diapedesis | 13/190 (7%) ICAM1, PPBP, ITGA5, CXCL5, SDC4, CCL3, GNAI2, MYL9, CLDN5, CCL3L3, IL1B, CXCL1, CXCL2 | 1/190 (1%) ITGB1 |
| TREM1 Signaling | 7/76 (9%) ICAM1, AKT1, NLRP12, ITGA5, IL1B, CD83, CCL3 | 2/76 (3%) ITGB1, NLRC3 |
| Canonical Pathways . | Downregulated genes . | Upregulated genes . |
|---|---|---|
| Integrin Signaling | 15/201 (7%) ITGA2B, MAP3K11, ITGA5, MYLK, ITGB3, MYL9, PARVB, AKT1, RHOB, CAPN1, ACTN4, CTTN, ARPC4, ACTN1, ITGB5 | 2/201 (1%) ITGB1, PPP1R12A |
| Granulocyte Adhesion and Diapedesis | 14/179 (8%) CSF3R, ICAM1, PPBP,ITGA5, CXCL5, SDC4, CCL3, ITGB3, GNAI2, CLDN5, CCL3L3, IL1B, CXCL1, CXCL2 | 1/179 (1%) ITGB1 |
| Signaling by Rho Family GTPases | 13/236 (6%) SEPT5, MAP3K11, ITGA5, MYLK, GNAZ, CDC42EP2, GNAI2, MYL9, GNG11, GNA15, RHOB, GNB2, ARPC4 | 3/236 (1%) ITGB1, DIAPH3, PPP1R12A |
| Agranulocyte Adhesion and Diapedesis | 13/190 (7%) ICAM1, PPBP, ITGA5, CXCL5, SDC4, CCL3, GNAI2, MYL9, CLDN5, CCL3L3, IL1B, CXCL1, CXCL2 | 1/190 (1%) ITGB1 |
| TREM1 Signaling | 7/76 (9%) ICAM1, AKT1, NLRP12, ITGA5, IL1B, CD83, CCL3 | 2/76 (3%) ITGB1, NLRC3 |
Differentially expressed genes post-ECP between responders and non-responders
Duvic:Therakos: Research Funding. Ni:Therakos: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal