Abstract
Background: Despite advances, the majority of patients (pts) with acute myeloid leukemia (AML) will die of their disease. Current genetics-based risk classification schemes inadequately predict outcome. We sought to determine if a novel methylation-based biomarker could enhance risk stratification in AML.
Methods: Using a novel methylation assay (xMELP) validated for the clinical laboratory, we performed DNA methylation assessment at 17 previously identified prognostic loci to calculate a summary methylation statistic (M-score, Wertheim et al. Clin Chem 2015) for 166 pts with de novo AML treated at the University of Pennsylvania (UPENN). Targeted next-generation sequencing (NGS) of 33 hematologic malignancy-associated genes was performed for a 136 pt subset. The association of M-score with other prognostic variables and outcome [complete remission (CR) and overall survival (OS)] was evaluated. Median follow-up was 68.1 mos (range 1.4-150.2) among 38 survivors and 10.5 mos (range 0.1-95.2) among those (n=128) deceased. The optimal M-score for identifying groups with differing OS in the total UPENN cohort defined a binary classifier. The classifier was validated in UPENN subgroups and in a separate ECOG ACRIN E1900 trial cohort.
Results: The mean and median M-score for the UPENN cohort was 92.3 (95% confidence interval [CI], 87.4-97.2) and 91.4 (range, 30.8-97.3), respectively; the M-score was not significantly associated with age, gender, specimen type, blast percent, NPM1, or FLT3-ITD. Patients with favorable cytogenetics had a lower mean M-score than those with non-favorable cytogenetics; there was no difference between intermediate and unfavorable cytogenetic groups. Univariate analyses demonstrated that a 10-unit increase in M-score was associated with a 10% increase in the hazard of death (HR 1.1; P<.0001) and a 20% increase in the odds of failing to achieve CR (OR 1.2, P<.0001). In multivariable Cox analysis, higher M-score (HR 1.1, P=.011) and older age (P=.001) were significantly associated with increased hazard of death, while NPM1+/FLT3-ITD-status (P=.031) was associated with decreased hazard of death. In a multivariable logistic analysis, higher M-score (HR, 1.1, P=.034) and older age (P=.007) were associated with increased odds of failing to achieve CR, while favorable cytogenetics (P=.030) was associated with achievement of CR.
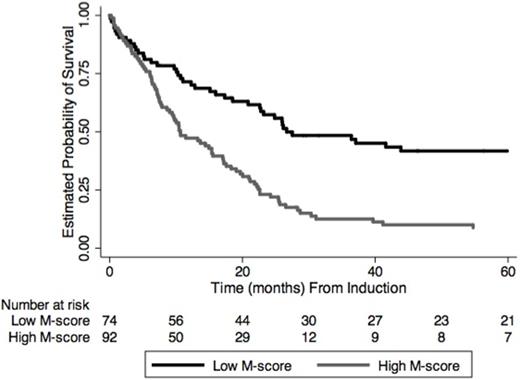
Based on the maximization of the log-rank statistic, the optimal M-score cutpoint was 86, which defined a binary risk classifier (hazard of death for high vs low M-score: unadjusted HR 2.5, P<.0001; adjusted HR 1.9, P=.003). Median OS was 26.6 vs. 10.6 mos for low and high M-score groups (Figure). The CR rates for low and high M-score groups were 84% and 61%, respectively. The performance of the M-score classifier was confirmed in pts ≤ 60 yrs with intermediate cytogenetics (log-rank P=.001, OS in high vs low M-score groups: 36.4 vs 14.0 mos) and among pts who achieved CR with initial therapy (log-rank P<.00001, OS 43.9 vs 17.2 mos). The M-score classifier identified groups with different outcome regardless of allogeneic transplant.
The M-score classifier was validated in the E1900 cohort (n=383). The mean and median M-scores were similar to the UPENN cohort and M-score was significantly associated with OS on multivariable analysis (P<0.0001). The M-score classifier identified E1900 subgroups with different OS (log-rank P<.00001; OS 29.5 vs 12.6 mos), a finding confirmed in the intermediate cytogenetics group, in those aged <50 and ≥50 yrs, and among pts assigned to both standard and high dose daunorubicin. Notably, high dose daunorubicin benefited patients with high M-scores (P=.001), but not those with low M-scores (P=.328).
We also evaluated the prognostic value of the M-score in the context of an extended molecular profile. Random forest analysis of the UPENN cohort showed that M-score and age are the most robust predictors of OS, while a subset of recurrent mutations (FLT3-ITD, NPM1, IDH2, TET2, TP53, NRAS, CEBPA) also contribute to prognosis, but to a lesser degree.
Conclusion: The M-score and associated classifier represent promising tools for clinical decision-making in AML and deserve further investigation. The prognostic value of the M-score may be superior to mutational analysis. Optimal methods for integration of M-score, clinical and genetic information are being defined.
Loren:Merck: Research Funding. Levine:Foundation Medicine: Consultancy; CTI BioPharma: Membership on an entity's Board of Directors or advisory committees; Loxo Oncology: Membership on an entity's Board of Directors or advisory committees.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal