Key Points
Somatic mutations alter nuclear distribution and association of CHD2 with actively transcribed genes in CLL.
CHD2 is the most frequently mutated CLL driver in the IGHV-mutated prognostic subgroup.
Abstract
Great progress has recently been achieved in the understanding of the genomic alterations driving chronic lymphocytic leukemia (CLL). Nevertheless, the specific molecular mechanisms governing chromatin remodeling in CLL are unknown. Here we report the genetic and functional characterization of somatic mutations affecting the chromatin remodeler CHD2, one of the most frequently mutated genes in CLL (5.3%) and in monoclonal B lymphocytosis (MBL, 7%), a B-cell expansion that can evolve to CLL. Most of the mutations affecting CHD2, identified by whole-exome sequencing of 456 CLL and 43 MBL patients, are either truncating or affect conserved residues in functional domains, thus supporting a putative role for CHD2 as a tumor suppressor gene. CHD2 mutants show altered nuclear distribution, and a chromodomain helicase DNA binding protein 2 (CHD2) mutant affected in its DNA-binding domain exhibits defective association with active chromatin. Clinicobiological analyses show that most CLL patients carrying CHD2 mutations also present mutated immunoglobulin heavy chain variable region genes (IGHVs), being the most frequently mutated gene in this prognostic subgroup. This is the first study providing functional evidence supporting CHD2 as a cancer driver and opens the way to further studies of the role of this chromatin remodeler in CLL.
Introduction
Chronic lymphocytic leukemia (CLL) is the most prevalent B-cell neoplasm in Western adults. CLL shows a remarkably heterogeneous course, with some patients presenting an indolent disease and normal life span, whereas others suffer from a dismal prognosis.1-3 In general, disease outcome correlates with the immunoglobulin heavy chain variable region genes (IGHV) mutational status of tumor lymphocytes,4,5 but also with other biological features including chromosomal aberrations and the expression of CLL-specific biomarkers.5-8
Recently, whole-genome sequencing–based reports have uncovered NOTCH1, MYD88, and XPO1 as recurrently mutated CLL drivers.9 Additionally, whole-exome sequencing approaches have identified the splicing factor SF3B110-13 and the shelterin complex member POT114 as CLL drivers associated with unfavorable prognosis. Also, mutations in sucrose isomaltase have been proposed to play a role in the CLL leukemogenic metabolic switch.15 Moreover, clonal evolution events in CLL have been delineated, pointing to the presence of subclonal driver mutations as a relevant prognostic factor in this disease.16 Notably, mutations in many of the previously described driver genes, as well as in BRAF, EGR2, and NFKBIE, can be detected in pluripotent hematopoietic progenitor cells, indicating that founding CLL mutations appear as very early evolutionary events.17 These genomic studies have been extended by comprehensive transcriptome profiling18 as well as by DNA methylome analyses.19 Despite this wealth of recent information, the need for the functional characterization and clinical description of many genetic alterations in CLL still remains, even for some of the most recurrently mutated genes. This is the case of the chromodomain helicase DNA binding protein 2 (CHD2), one of the most frequently mutated genes in CLL.10,16 The functional roles of CHD2, a member of the CHD family of SNF2 adenosine triphosphate (ATP)–dependent chromatin remodelers, are still poorly defined. Homozygous Chd2 mutant mice show embryonic growth delay and perinatal lethality, and heterozygous animals undergo reduced neonatal viability and decreased life span.20,21 Notably, most Chd2 heterozygous mutants die because of multiple lymphomas and lymphoid hyperplasias,21 supporting CHD2 as a candidate driver gene in hematologic malignancies.
Here we describe the CHD2 mutations identified in a cohort of 456 CLL patients and 43 cases of monoclonal B lymphocytosis (MBL) CLL type.22 We also present functional, transcriptomic, and epigenomic analyses showing that CHD2 mutations influence chromatin remodeling processes in CLL. Finally, we analyze the clinicobiological features of patients with CHD2 mutations.
Materials and methods
Patients
Four hundred and fifty-six patients with CLL and 43 with MBL according to World Health Organization criteria were included in the study. Tumor samples were always collected before the administration of any treatment. The main clinical characteristics of the patients are detailed in supplemental Table 1 (see the Blood Web site). All patients gave informed consent for their participation in the study following the International Cancer Consortium guidelines.23
Statistical analysis
The 2-tailed Fisher’s exact test was used to correlate clinicobiological variables according to CHD2 mutations. Time to treatment (time from diagnosis to first treatment) and overall survival (time from diagnosis to the date of death or to the date of last follow-up for surviving patients) curves were compared by the log-rank test. All the analyses were conducted with SPSS 20 (www.ibm.com).
DNA constructs and site-directed mutagenesis
Full-length CHD2 transcript variant 1, cloned with an in-frame C-terminal Myc-DDK tag in the pCMV6-Entry vector, was from OriGene technologies. CHD2 variants were generated by site-directed mutagenesis using the QuickChange XL site-directed mutagenesis kit (Stratagene) and the oligonucleotides 5′-GGGAGTGGATGAAGCCCTTCGGTTGAAGAATGATG-3′ and 5′-CATCATTCTTCAACCGAAGGGCTTCATCCACTCCC-3′ for the H620L mutant, 5 5′-GGTTCATCAAGGCTTATAAGAAGTTAGGTCTCCCTCTTGAA-3′ and 5′-TTCAAGAGGGAGACCTAACTTCTTATAAGCCTTGATGAACC-3′ for the F1146L form, and 5′-TGGAAGATGATTCTCGCCTGTTCCTGGGGATTTAT-3′ and 5′-ATAAATCCCCAGGAACAGGCGAGAATCATCTTCCA-3′ for the L1270F form. All constructs were sequenced and validated before experimental use.
Cell culture, transfection, and nucleofection
COS-7 and HEK293T cells were maintained in Dulbecco’s modified Eagle medium (Gibco), supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin-glutamine (Gibco). Transfections were carried out in cells seeded onto gelatinized coverslips and using lipofectamine plus or lipofectamine 3000 (Invitrogen), following the manufacturer’s instructions. For nucleofection, the CLL cell line MEC-1 was cultured at a density of 1 × 106 cells per mL in Iscove's modified Dulbecco's medium (Gibco) supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin-glutamine (Gibco) 1 day before. For each nucleofection reaction, 3 µg of DNA was used to transfect 3 × 106 cells in 100 µL nucleocuvettes employing the nucleofection kit V (Lonza) and the program U-016 in a Nucleofector 2b device (Lonza). Then, cells were incubated at 37°C in 500 μL of warm RPMI for recovery and plated after 10 minutes in 12-well plates containing prewarmed complete Iscove's modified Dulbecco's medium.
Confocal immunofluorescence microscopy
Transfected cells were washed with phosphate-buffered saline (PBS) and fixed for 15 minutes at room temperature in 4% p-formaldehyde-buffered solution. Cells were permeabilized with 0.5% triton X-100 and blocked with 15% goat serum for 1 hour at room temperature, followed by an overnight coincubation with rabbit anti-DYKDDDDK (anti-FLAG) antibody (1:800; Cell Signaling) and either mouse anti-acetyl histone H4 (1:250; Millipore), mouse anti-H3K4me3 (1:250; Active Motif), or mouse anti-SC35 (1 μg/mL; Abcam). Immunofluorescence was detected using Alexa Fluor 488 anti-rabbit and Alexa Fluor 546 anti-mouse secondary antibodies. The coverslips were mounted on slides using 4,6 diamidino-2-phenylindole–containing Vectashield ProLong Gold antifade agent (Molecular Probes) and imaged in a Leica TCS-SP2-AOBS confocal microscope (Leica Microsystems). Images were processed using Image J software. Colocalization analyses and quantification were performed using the colocalization threshold plug-in developed by Dr. T. Collins (Wright Cell Imaging Facility, Toronto, ON, Canada). Quantification of colocalization was performed following the Costes method for Pearson coefficient minimization in order to finally estimate the background-thresholded Manders' colocalization coefficient.24 A minimum of 10 cells were imaged to estimate mean colocalization values per sample. For nucleofection analyses, MEC-1 cells were transferred to 12-well plates 24 hours after nucleofection and centrifuged over poly-l-lysine–coated glass slides placed in the bottom of the wells. Cells were allowed to attach to the coated slides for 2 hours at 37°C and then processed for immunofluorescence-based confocal microscopy as described previously.
Immunoprecipitation and immunoblotting assays
HEK293T cells cultured in 100-mm dishes were transfected with an empty vector (mock control) or the pCMV-6-Entry plasmid harboring CHD2wt, CHD2H620L, or CHD2L1270F. Twenty-four hours after transfection, cells were washed with cold PBS, collected by trypsinization, and resuspended in coimmunoprecipitation (co-IP) lysis buffer composed of 50 mM tris(hydroxymethyl)aminomethane (Tris)–HCl pH 8, 150 mM NaCl, 1.5 mM MgCl2, 5 mM EDTA, 1 mM dithiothreitol (DTT), 1% NP-40, 20% glycerol, and protease inhibitor cocktail (EDTA-free; Roche Applied Science). Cells were subjected to several freeze-thaw cycles, followed by 5 sonication pulses, and centrifuged at maximum speed for 30 minutes in a refrigerated microcentrifuge. The soluble fractions were incubated overnight at 4°C with 10 µL of mouse anti-FLAG antibody (Sigma) followed by 2 hours incubation with protein G magnetic dynabeads (Life Technologies). Then, the beads were washed with washing buffer (50 mM Tris-HCl pH 8, 150 mM NaCl, 1.5 mM MgCl2, 5 mM EDTA, 1 mM DTT, and 0.1% NP-40) and eluted by boiling them in 50 µL of sodium dodecyl sulfate–polyacrylamide gel electrophoresis loading buffer. Finally, 10 µL of the samples were loaded onto 4% to 20% precast polyacrylamide gels (BioRad) and electrophoresed for western blot analysis. The proteins were electrotransferred onto polyvinylidene difluoride membranes, which were blocked afterward with 5% bovine serum albumin or 5% nonfat dried milk in Tris-buffered saline with 0.05% Tween-20 (TBST), and incubated with either rabbit or mouse monoclonal anti-FLAG antibody (1:1000; Cell Signaling), mouse anti-acetyl histone H4 (Lys5/8/12/16; 1:2000; Millipore), or mouse anti-histone H3K4me3 (1:225; Active Motif). After washing with TBST, membranes were incubated with horseradish peroxidase–conjugated goat anti-rabbit or anti-mouse IgG (Thermo Scientific) at a 1:104 dilution in 2.5% milk in TBST and developed with the Luminata Forte Western HRP substrate (Millipore).
Subcellular fractionation
HEK293T cells were cultured in 6-well plates and transfected with an empty vector (mock control) or the pCMV-6-Entry plasmid harboring CHD2wt, CHD2H620L, or CHD2L1270F. After 24 hours, cells were washed with PBS, trypsinized, and centrifuged. For formaldehyde crosslinking, pelleted cells were resuspended in 0.8 mL of PBS containing 0.2% formaldehyde and incubated for 5 minutes at room temperature. Afterward, excess formaldehyde was quenched with 100 mM glycine for 5 minutes followed by a 5-minute centrifugation step at 300g. After removal of the supernatant, cells were resuspended in outer-membrane lysis buffer (10 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid pH 7.9, 10 mM KCl, 0.1% NP-40, 1 mM MgCl2, 1 mM DTT, and protease inhibitor cocktail) and incubated for 10 minutes on ice. The cytosolic fraction was then collected after 5 minutes of centrifugation at 1000g on a refrigerated microcentrifuge. The pelleted fraction was washed with the same buffer, centrifuged, and incubated in 25 µL of nuclear lysis buffer (10 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid pH 7.9, 420 mM KCl, 1% NP-40, 0.2% sodium dodecyl sulfate, 1 mM MgCl2, 1 mM DTT, 25% glycerol, and protease inhibitor cocktail) for 15 minutes on ice. Afterward, the lysated nuclei were sonicated and centrifuged at maximum speed for 5 minutes. Finally, the nuclear fraction (supernatant) was diluted with additional 25 µL of nuclear lysis buffer containing neither detergents nor KCl. For immunobloting, 30 µg of each fraction were loaded onto 4% to 20% precast polyacrylamide gels and processed as described previously. The proteins calreticulin and histone H2B were respectively used as cytoplasmic and nuclear markers for quality control assessment of the subcellular fractionation process employing mouse anti-calreticulin (Abcam) and rabbit anti-histone H2B (Upstate Biotechnology) antibodies.
Whole-transcriptome analysis
Two groups of patients were selected to search for CHD2-related expression patterns (supplemental Table 2). The CHD2mut group includes tumors with CHD2 mutation and classified as IGHV mutated. The control group contains the same number of IGHV-mutated tumors with no mutation in CHD2 as assessed by whole-exome sequencing. All patients in both groups were male. We extracted expression data from Affymetrix Human Genome Array U219 array plates hybridized with high quality RNA from CHD2mut and control samples. We used the Robust Multiarray Average in the Expression Console software (Affymetrix) to generate summarized expression values from the raw microarray values. The tabulated data were then fed into the Gene Set Enrichment Analysis (GSEA) algorithm through the R interface provided by the Broad Institute.25 We ran 100 permutations of the algorithm using the msigdb.v3.0.symbols.gmt Molecular Signatures Database26 and default parameters. We set the significance threshold at the 0.25 false discovery rate q value. Expression array data are stored at the European Genome-Phenome Archive (http://www.ebi.ac.uk/ega/), which is hosted by the European Bioinformatics Institute, under accession number EGAD00010000472.
Results
Structural impact of CHD2 mutations detected in CLL and MBL patients
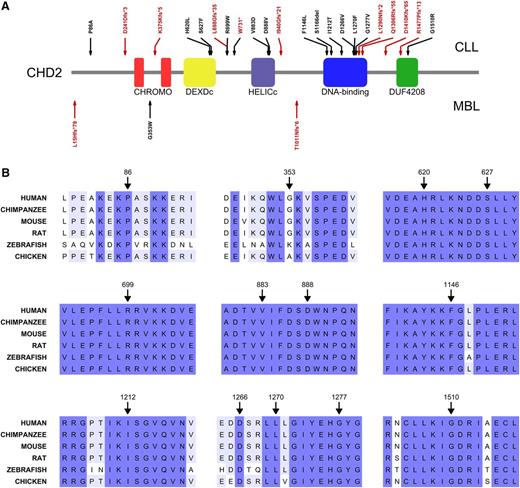
CHD2 is a complex multidomain protein composed of N-terminal tandem chromodomains (chromatin organization modifier domain), followed by a DEXDc (Dead-like helicase superfamily) domain and a HELICc (helicase superfamily C-terminal) domain, both spanning the SNF2 (SNF2 family N-terminal) domain. CHD2 also contains a putative DBD (DNA binding domain) and a C-terminal domain of unknown function (DUF4208) (Figure 1A). Somatic mutations in CHD2 were identified in 24 out of 456 CLL cases (5.3%) as well as in 3 out of 43 (7%) MBL cases. The methods for whole-genome, whole-exome, and mutational analysis have been previously published10,14 and are detailed in the supplemental Methods. A comprehensive genomic analysis of both series will be reported elsewhere (ICGC CLL Consortium, unpublished data). Importantly, many of the CHD2 mutations identified are predicted to result in truncated protein forms (either through frameshifts, altered splicing sites, or premature stop codons), reaching up to 46% of the mutants found in CLL. All SNVs affect highly conserved positions, with 11 of these mutated residues located inside functional domains (Figure 1 and Table 1), thus suggesting profound biological influence of the CHD2 alterations in both B-cell neoplasms. All missense mutations were predicted to be deleterious by CONDEL, a method that assesses the outcome of SNVs by combining 5 predictive tools,27 with the only exception of P86A in CLL and G353W in MBL. Sequence alignment of yeast Chd1 and human CHD1, CHD2, and CHD6-9 shows that the putative CHD2 DBD can be delineated through sequence homology with the Chd1 DBD region (supplemental Figure 1). This CHD2 domain is mutated in >33% of the CLL cases, presenting 3 frameshift mutations (Figure 1) and 5 SNVs affecting highly conserved residues (supplemental Figure 1).
Structural impact of CHD2 mutations in CLL and MBL. (A) Schematic representation of CHD2 structure and distribution of the mutations identified in the cohorts of CLL (n = 456) and MBL (n = 43). Red arrows indicate frameshift mutations, splice site alterations, and premature stop codons; whereas black arrows represent single nucleotide variants (SNVs) and a single residue deletion. CHROMO, chromatin organization modifier domain; DEXDc, Dead-like helicase superfamily; DUF4208, domain of unknown function 4208; HELICc, helicase superfamily c-terminal. (B) Amino acid sequence alignments of the regions surrounding the identified SNVs show the high phylogenetic conservation of the affected residues.
Structural impact of CHD2 mutations in CLL and MBL. (A) Schematic representation of CHD2 structure and distribution of the mutations identified in the cohorts of CLL (n = 456) and MBL (n = 43). Red arrows indicate frameshift mutations, splice site alterations, and premature stop codons; whereas black arrows represent single nucleotide variants (SNVs) and a single residue deletion. CHROMO, chromatin organization modifier domain; DEXDc, Dead-like helicase superfamily; DUF4208, domain of unknown function 4208; HELICc, helicase superfamily c-terminal. (B) Amino acid sequence alignments of the regions surrounding the identified SNVs show the high phylogenetic conservation of the affected residues.
Somatic mutations affecting CHD2 in CLL and MBL patients
| Case . | Genomic . | Exon . | Mutation . | AF . | Type . | Prediction . | IGHV status . |
|---|---|---|---|---|---|---|---|
| CLL | |||||||
| 7 | 93552555 | 35-36 | — | 0.20-0.52 | Splicing | — | Mutated |
| 17 | 93467744 | 3 | p.P86A | 0.40-0.59 | Missense | Neutral | Unmutated |
| 43 | 93540558 | 30 | p.L1270F | 0.23-0.50 | Missense | Deleterious | Mutated |
| 175 | 93510647 | 17 | p.L698Gfs*35 | 0.32-0.57 | Deletion | — | Mutated |
| 181 | 93534730 | 27 | p.F1146L | 0.37-0.64 | Missense | Deleterious | Mutated |
| 185 | 93499738 | 16 | p.H620L | 0.39-0.64 | Missense | Deleterious | Mutated |
| 274 | 93522456 | 22 | p.I940Gfs*21 | 0.12-0.29 | Deletion | — | Mutated |
| 381 | 93514997 | 18 | p.W731* | 0.26-0.48 | Nonsense | — | Mutated |
| 569 | 93521534 | 21 | p.V883D | 0.38-0.55 | Missense | Deleterious | Mutated |
| 591 | 93521549 | 21 | p.D888V | 0.42-0.67 | Missense | Deleterious | Mutated |
| 709 | 93536130 | 28 | p.S1166del | 0.16-0.43 | Deletion | — | Mutated |
| 723 | 93545499 | 33 | p.D1410Kfs*65 | 0.35-0.58 | Deletion | — | Unmutated |
| 767 | 93540618 | 30 | p.L1290Nfs*2 | 0.30-0.50 | Deletion | — | Mutated |
| 788 | 93552392 | 35 | p.R1477Pfs*13 | 0.35-0.68 | Insertion | — | Mutated |
| 853 | 93485082 | 8 | p.D241Dfs*3 | 0.23-0.52 | Insertion | — | Unmutated |
| 880 | 93540545 | 30 | p.D1266V | 0.13-0.28 | Missense | Deleterious | Mutated |
| 1162 | 93540578 | 30 | p.G1277V | 0.09-0.46 | Missense | Deleterious | Mutated |
| 1177 | 93541759 | 31 | p.Q1306Rfs*55 | 0.16-0.70 | Deletion | — | Mutated |
| 1191 | 93540226 | 29 | p.I1212T | 0.29-0.63 | Missense | Deleterious | Mutated |
| 1192 | 93499759 | 16 | p.S627F | 0.14-0.43 | Missense | Deleterious | Unmutated |
| 1343 | 93487716 | 10 | p.K375Kfs*5 | 0.26-0.56 | Insertion | — | Mutated |
| 1424 | 93510649 | 17 | p.R699W | 0.31-0.64 | Missense | Deleterious | Mutated |
| 1431 | 93510649 | 17 | p.R699W | 0.46-0.77 | Missense | Deleterious | Unmutated |
| 1535 | 93552489 | 35 | p.G1510R | 0.21-0.65 | Missense | Deleterious | Mutated |
| MBL | |||||||
| 152 | 93444511 | 2 | p.L15Hfs*79 | 0.24-0.71 | Insertion | — | — |
| 190 | 93487649 | 10 | p.G353W | 0.09-0.48 | Missense | Neutral | — |
| 383 | 93524650 | 24 | p.T1011Nfs*6 | 0.19-0.44 | Insertion | — | — |
| Case . | Genomic . | Exon . | Mutation . | AF . | Type . | Prediction . | IGHV status . |
|---|---|---|---|---|---|---|---|
| CLL | |||||||
| 7 | 93552555 | 35-36 | — | 0.20-0.52 | Splicing | — | Mutated |
| 17 | 93467744 | 3 | p.P86A | 0.40-0.59 | Missense | Neutral | Unmutated |
| 43 | 93540558 | 30 | p.L1270F | 0.23-0.50 | Missense | Deleterious | Mutated |
| 175 | 93510647 | 17 | p.L698Gfs*35 | 0.32-0.57 | Deletion | — | Mutated |
| 181 | 93534730 | 27 | p.F1146L | 0.37-0.64 | Missense | Deleterious | Mutated |
| 185 | 93499738 | 16 | p.H620L | 0.39-0.64 | Missense | Deleterious | Mutated |
| 274 | 93522456 | 22 | p.I940Gfs*21 | 0.12-0.29 | Deletion | — | Mutated |
| 381 | 93514997 | 18 | p.W731* | 0.26-0.48 | Nonsense | — | Mutated |
| 569 | 93521534 | 21 | p.V883D | 0.38-0.55 | Missense | Deleterious | Mutated |
| 591 | 93521549 | 21 | p.D888V | 0.42-0.67 | Missense | Deleterious | Mutated |
| 709 | 93536130 | 28 | p.S1166del | 0.16-0.43 | Deletion | — | Mutated |
| 723 | 93545499 | 33 | p.D1410Kfs*65 | 0.35-0.58 | Deletion | — | Unmutated |
| 767 | 93540618 | 30 | p.L1290Nfs*2 | 0.30-0.50 | Deletion | — | Mutated |
| 788 | 93552392 | 35 | p.R1477Pfs*13 | 0.35-0.68 | Insertion | — | Mutated |
| 853 | 93485082 | 8 | p.D241Dfs*3 | 0.23-0.52 | Insertion | — | Unmutated |
| 880 | 93540545 | 30 | p.D1266V | 0.13-0.28 | Missense | Deleterious | Mutated |
| 1162 | 93540578 | 30 | p.G1277V | 0.09-0.46 | Missense | Deleterious | Mutated |
| 1177 | 93541759 | 31 | p.Q1306Rfs*55 | 0.16-0.70 | Deletion | — | Mutated |
| 1191 | 93540226 | 29 | p.I1212T | 0.29-0.63 | Missense | Deleterious | Mutated |
| 1192 | 93499759 | 16 | p.S627F | 0.14-0.43 | Missense | Deleterious | Unmutated |
| 1343 | 93487716 | 10 | p.K375Kfs*5 | 0.26-0.56 | Insertion | — | Mutated |
| 1424 | 93510649 | 17 | p.R699W | 0.31-0.64 | Missense | Deleterious | Mutated |
| 1431 | 93510649 | 17 | p.R699W | 0.46-0.77 | Missense | Deleterious | Unmutated |
| 1535 | 93552489 | 35 | p.G1510R | 0.21-0.65 | Missense | Deleterious | Mutated |
| MBL | |||||||
| 152 | 93444511 | 2 | p.L15Hfs*79 | 0.24-0.71 | Insertion | — | — |
| 190 | 93487649 | 10 | p.G353W | 0.09-0.48 | Missense | Neutral | — |
| 383 | 93524650 | 24 | p.T1011Nfs*6 | 0.19-0.44 | Insertion | — | — |
AF, allelic frequency (95% CI); CI, confidence interval.
An independent whole-exome–based CLL study reported CHD2 to be recurrently mutated in 3.75% of a cohort of 160 patients.16 Half of the mutations identified in this work were truncating, in good agreement with our data. A search in the COSMIC database28 shows that CHD2 is also frequently mutated in some solid tumors, mainly large intestine, urinary tract, and endometrium carcinomas. This information supports CHD2 as a novel gene involved in cancer development and points to a role for CHD2 as a CLL driver gene, most likely through protein loss of function.
Nuclear distribution of CHD2 mutants
CHD chromatin remodelers modulate the expression of many different genes.29,30 According to structural features, the CHD family is divided in 3 subfamilies, with subfamily I composed of CHD2 and its closest homolog CHD1, which interacts with the trimethylated histone H3 lysine 4 (H3K4me3) mark and is associated with transcriptional activation in different organisms.29-31 CHD2 carries the CHD1 residues responsible for H3K4me3 binding, and recent works have shown an almost exclusive preference of CHD2 for active promoter transcription start sites, associated with active chromatin marks.32 In contrast, other CHD family members such as CHD3 and CHD4 have been described as expression repressors.33,34 These data, together with the reported role of CHD2 in myogenic differentiation,35 suggest that the putative effects of CHD2 mutation in CLL could be related to the regulation of active gene transcription. We therefore set out to assess the distribution of wild-type and mutated CHD2 proteins in the cell nucleus and their association with transcriptionally active chromatin.
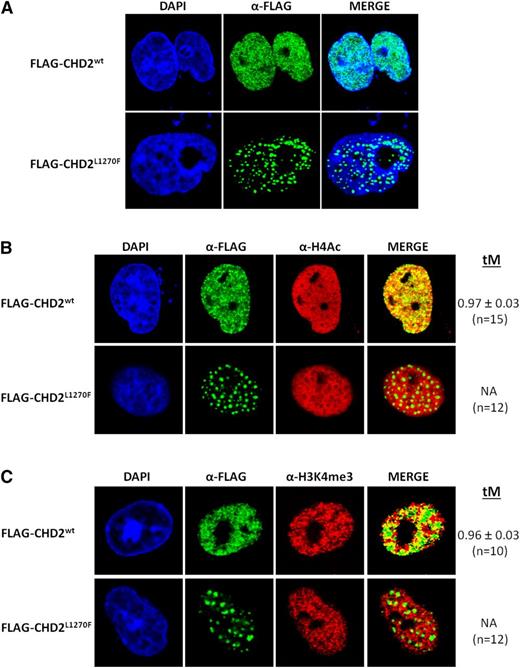
Fluorescence-based confocal microscopy imaging analysis of transfected COS-7 cells shows a stark contrast between the nuclear distribution of recombinant human CHD2wt and the CLL-associated mutant form CHD2L1270F (Figure 2A). CHD2wt shows the expected homogeneous and finely granulated distribution throughout the nucleus and exclusion from the nucleoli, in good agreement with the distribution previously observed in murine cells.35 On the contrary, the modification of a conserved residue in the DBD seems to drive to the accumulation of CHD2L1270F in large discrete punctated bodies. A strikingly similar punctated distribution has been previously described in a study performed with different shortened and truncated forms of CHD1.36
Fluorescence-based confocal imaging analysis of CHD2 mutations in COS-7 cells. (A) Nuclear distribution of CHD2wt and CHD2L1270F. Recombinant CHD2wt shows a finely granulated homogeneous distribution within the cell nucleus, whereas the CHD2L1270F mutation induces a remarkably speckled distribution of the protein in large punctated bodies. (B-C) Association of CHD2wt and CHD2L1270F with active chromatin. CHD2wt shows a strong colocalization with the transcriptionally active chromatin markers H4Ac and H3K4me3. Strong functional impact can be predicted from the complete loss of colocalization of the mutant form CHD2L1270F with the gene-expressing chromatin regions. tM is expressed as mean ± standard deviation. n, number of nuclei analyzed per sample.
Fluorescence-based confocal imaging analysis of CHD2 mutations in COS-7 cells. (A) Nuclear distribution of CHD2wt and CHD2L1270F. Recombinant CHD2wt shows a finely granulated homogeneous distribution within the cell nucleus, whereas the CHD2L1270F mutation induces a remarkably speckled distribution of the protein in large punctated bodies. (B-C) Association of CHD2wt and CHD2L1270F with active chromatin. CHD2wt shows a strong colocalization with the transcriptionally active chromatin markers H4Ac and H3K4me3. Strong functional impact can be predicted from the complete loss of colocalization of the mutant form CHD2L1270F with the gene-expressing chromatin regions. tM is expressed as mean ± standard deviation. n, number of nuclei analyzed per sample.
In the case of 2 additional CHD2 mutants, which affect either another conserved residue in the DBD (CHD2F1146L) or the DEAH box of the helicase domain (CHD2H620L), the change from a fine homogeneous distribution to a more punctated appearance was moderate but still present and more evident for the DBD mutant (supplemental Figure 2A). This heterogeneity is also concordant with that observed before for the expression of CHD1 mutants.36 Notably, both CHD2H620L and CHD2F1146L showed lower expression levels when compared with CHD2wt and CHD2L1270F. In fact, CHD2F1146L was rarely expressed, and often at very reduced levels (data not shown). Here, putative loss of function could be enhanced by the generation of unstable gene products.
Colocalization tests confirmed the strong association of CHD2wt with transcriptionally active chromatin markers, namely acetyl histone H4 (H4Ac) and H3K4me3, showing colocalization values ∼96% according to the thresholded Manders' colocalization coefficients obtained (tM; Figure 2B-C). This result supports the proposed link between CHD2 and transcriptional activation processes. Additional experiments showed that the colocalization with both H4Ac and H3K4me3 marks was abolished for the CHD2L1270F nuclear bodies, likely entailing strong functional consequences in the cells harboring this mutant form. No significant loss of colocalization was detected for the CHD2H620L and CHD2F1146L mutants despite the apparent defects observed for their expression and nuclear distribution (supplemental Figure 2B-C), showing the Manders' coefficient values similar to those of CHD2. As suggested earlier, haploinsufficiency could be also accounting for functional defects in these cases.
Importantly, both the nuclear distribution and the colocalization properties of CHD2wt, CHD2H620L, and CHD2L1270F with the H4Ac marks were further validated in the MEC-1 CLL cell line (supplemental Figure 3). Thus, the high tM values observed for CHD2wt and CHD2H620L colocalization with the histone H4Ac drop dramatically (tM = 0.31) for CHD2L1270F, thus confirming the observed functional defect of this mutant in the nuclei of CLL cells. On the other hand, physical and functional association of CHD1 with pre–messenger RNA splicing factors has been documented before31 and the nuclear bodies of CHD2L1270F resemble known nuclear speckles harboring spliceosome components.37 The confocal studies reflect no significant colocalization of any of the proteins with SC-35, a nuclear speckle marker (supplemental Figure 4).
Interaction of CHD2 mutants with active chromatin marks
To further evaluate the association of wild-type and mutant forms of CHD2 with active chromatin marks, co-IP experiments were performed upon expression of recombinant CHD2wt, CHD2H620L, and CHD2L1270F in HEK293T cells. Interestingly, the 3 different CHD2 variants tested displayed the ability to interact with both H3K4me3 and H4Ac specifically, in contrast with the lack of pulled-out protein observed in the co-IP performed with an empty-vector control (supplemental Figure 5A). These results suggest that the null colocalization observed between the CHD2L1270F mutant and actively transcribed chromatin is caused by the inability of the DBD-mutant protein to directly interact with the DNA molecule, rather than with the modified histones, an interaction that has been shown to depend on the chromodomains.33
The defective interaction of CHD2L1270F with its target nuclear structures was also supported by the behavior observed for this mutant in subcellular fractionation experiments. Thus, whereas both CHD2wt and CHD2H620L present an almost exclusively nuclear distribution upon fractionation, a very important portion of the total CHD2L1270F can be detected in the fraction corresponding to the cytoplasmic compartment (supplemental Figure 5B). Because confocal microscopy imaging showed that all the protein forms under study are exclusively nuclear, the presence of CHD2L1270F in the cytoplasmic fraction can be only explained by diffusion of the protein after soft permeabilization of the nuclear membrane during the extraction of the cytoplasmic fraction. Therefore, although the wild-type protein and the DEAH box mutant remain associated to chromatin in the nuclear compartment, the CHD2L1270F mutant would be eventually released to the cytoplasm, likely because of reduced affinity for its nuclear target. The exclusively nuclear localization of CHD2L1270F in vivo was corroborated by formaldehyde crosslinking experiments prior to subcellular fractionation, which efficiently blocked the diffusion of the mutant form to the cytoplasmic compartment (supplemental Figure 5B).
Transcriptome and methylome analysis of CHD2-mutated cells from CLL patients
To obtain a comprehensive picture of the functional consequences of CHD2 mutations, we performed transcriptome and methylome analysis in cells from CLL patients. Data obtained from whole-genome gene expression arrays performed with 7 CHD2-mutated and 7 CHD2-unmutated patients, all of them with an IGHV-mutated status, were subjected to GSEA. Several functional pathways were found to be significantly enriched (false discovery rate <0.25) in the CHD2-mutated patients (supplemental Table 3). Importantly, some of the top-ranked pathways are associated with DNA modification processes (DNA-dependent ATPase activity, DNA helicase activity, and anaphase-promoting complex phosphorylation), and core enrichment in these pathways suggests that CHD2 could be modulating several chromatin remodeling processes, even through other DNA modifiers. Moreover, the identification of significant enrichment for genes belonging to the phosphatidylinositol-4, 5-bisphosphate 3-kinase (PI3K) pathway is remarkable in the context of B-cell signaling38 and, hence, possibly relevant for CLL development. Notably, some of the pathways identified in this transcriptomic analysis of CLL samples are related to brain physiology, thus establishing a putative link between CHD2 mutations and the documented role of pathogenic forms of this protein in different encephalopathies and neurologic disorders.39,40
No differences were observed for CHD2 gene methylation levels between mutated and wild-type CHD2 in CLL patients (supplemental Figure 6A). Global cytosine guanine dinucleotide methylation analysis uncovered >400 regions with differential methylation levels in CHD2-mutated patients (supplemental Figure 6B and supplemental Figure 7). The involvement of CHD2 in gene expression modulation is supported by significant functional annotation clustering of 25 transcription regulation genes and 6 negative regulation factors after gene ontology enrichment analysis of the hypomethylated and hypermethylated hits, respectively (supplemental Table 4). In good agreement with the GSEA, significant enrichment was also identified for hypomethylated genes associated to neuronal development and physiology. Notably, DLEU2, a frequently deleted gene in CLL,41 shows a significantly hypermethylated island in CHD2-mutated patients.
Clinical features of CLL patients with CHD2 mutations
The clinicobiological features of patients presenting CHD2 mutations were compared with those of the rest of the CLL cohort (Table 2). A close look at the biological and clinical parameters of the CLL patients shows a more frequently low CD49d expression in patients with CHD2 mutations, in the absence of other significant relationships. Despite the fact that 75% of CHD2-mutated patients present a mutated IGHV status, the association between CHD2 and IGHV mutation does not reach statistical significance, probably hampered by the low number of CHD2-mutated cases. Nevertheless, CHD2 is the most frequently mutated gene among all IGHV-mutated patients in our CLL cohort, involving 6.4% of all the cases. The high incidence of IGHV-mutated cases carrying CHD2 mutations prompted us to reassess the clinical features in the subgroup of 282 analyzed patients with an IGHV-mutated status (supplemental Table 5). No significant differences between the 2 groups emerged, although the results could again be affected by the relatively low number of cases with CHD2 mutations.
Clinical characteristics at sampling of the patients according to CHD2
| Parameter . | Category . | Unmutated (n = 432) . | Mutated (n = 24) . | P . |
|---|---|---|---|---|
| Gender | Male (%) | 260 (60) | 18 (75) | |
| Age (y), median (range) | 61 (18-93) | 64 (36-79) | ||
| A | 367 (86%) | 24 (100%) | ||
| Binet stage | B | 48 (11%) | 0 (0%) | |
| C | 11 (3%) | 0 (0%) | ||
| 0 | 274 (64%) | 19 (79%) | ||
| Rai stage | I-II | 136 (32%) | 5 (21%) | |
| III-IV | 15 (4%) | 0 (0%) | ||
| Lymphocytes (×109/L), median (range) | 11.1 (1.1-203) | 13.2 (5.9-80) | ||
| Hemoglobin (g/L), median (range) | 141 (45-177) | 144 (117-175) | ||
| Platelets (×109/L), median (range) | 201 (7-791) | 206 (101-301) | ||
| β2 microglobulin | UNV | 92 (21%) | 3 (12%) | |
| LDH | UNV | 32 (7%) | 2 (8%) | |
| IGHV | Mutated | 264 (62%) | 18 (75%) | |
| CD38 | <30% | 327 (76%) | 21 (88%) | |
| ZAP-70 | <20% | 323 (75%) | 20 (83%) | |
| CD49d | <40% | 334 (78%) | 22 (96%) | .038 |
| Genetics | Del(13q) | 215 (50%) | 8 (33%) | |
| Trisomy 12 | 63 (15%) | 5 (21%) | ||
| Del(11q) | 42 (10%) | 3 (12%) | ||
| Del(17p) | 15 (4%) | 2 (8%) | ||
| NOTCH1 | Mutated | 54 (13%) | 3 (13%) | |
| SF3B1 | Mutated | 40 (9%) | 0 (0%) | |
| MYD88 | Mutated | 17 (4%) | 1 (4%) | |
| TP53 | Mutated | 14 (3%) | 1 (4%) | |
| Treated | 184 (44%) | 11 (48%) | ||
| 5-y TTT (95% CI) | A and B | 53% (44%-62%) | 62% (28%-96%) | |
| 5-y OS (95% CI) | All | 79% (72%-86%) | 70% (46%-94%) |
| Parameter . | Category . | Unmutated (n = 432) . | Mutated (n = 24) . | P . |
|---|---|---|---|---|
| Gender | Male (%) | 260 (60) | 18 (75) | |
| Age (y), median (range) | 61 (18-93) | 64 (36-79) | ||
| A | 367 (86%) | 24 (100%) | ||
| Binet stage | B | 48 (11%) | 0 (0%) | |
| C | 11 (3%) | 0 (0%) | ||
| 0 | 274 (64%) | 19 (79%) | ||
| Rai stage | I-II | 136 (32%) | 5 (21%) | |
| III-IV | 15 (4%) | 0 (0%) | ||
| Lymphocytes (×109/L), median (range) | 11.1 (1.1-203) | 13.2 (5.9-80) | ||
| Hemoglobin (g/L), median (range) | 141 (45-177) | 144 (117-175) | ||
| Platelets (×109/L), median (range) | 201 (7-791) | 206 (101-301) | ||
| β2 microglobulin | UNV | 92 (21%) | 3 (12%) | |
| LDH | UNV | 32 (7%) | 2 (8%) | |
| IGHV | Mutated | 264 (62%) | 18 (75%) | |
| CD38 | <30% | 327 (76%) | 21 (88%) | |
| ZAP-70 | <20% | 323 (75%) | 20 (83%) | |
| CD49d | <40% | 334 (78%) | 22 (96%) | .038 |
| Genetics | Del(13q) | 215 (50%) | 8 (33%) | |
| Trisomy 12 | 63 (15%) | 5 (21%) | ||
| Del(11q) | 42 (10%) | 3 (12%) | ||
| Del(17p) | 15 (4%) | 2 (8%) | ||
| NOTCH1 | Mutated | 54 (13%) | 3 (13%) | |
| SF3B1 | Mutated | 40 (9%) | 0 (0%) | |
| MYD88 | Mutated | 17 (4%) | 1 (4%) | |
| TP53 | Mutated | 14 (3%) | 1 (4%) | |
| Treated | 184 (44%) | 11 (48%) | ||
| 5-y TTT (95% CI) | A and B | 53% (44%-62%) | 62% (28%-96%) | |
| 5-y OS (95% CI) | All | 79% (72%-86%) | 70% (46%-94%) |
LDH, lactate dehydrogenase; OS, overall survival; TTT, time to treatment; UNV, upper normal value; ZAP-70, zeta-chain-associated protein kinase 70kDa.
Discussion
A growing body of evidence shows that chromatin modification pathways are among the most frequently mutated in cancer.42 Notwithstanding that the delineation of CLL methylomes has been achieved,19 the molecular mechanisms modulating chromatin remodeling in CLL are still poorly understood. CHD2 was first identified as a frequently mutated CLL gene by whole-exome analysis of 105 CLL patients.10 An independent study confirmed the mutational recurrence of CHD2 and proposed that this gene, together with other mutated DNA modifiers and epigenetic factors, may support chromatin modification in CLL.16
To delve into the molecular and clinical impact of CHD2 mutations in leukemogenesis, we conducted further whole-exome and whole-genome sequencing analyses to interrogate 456 CLL and 43 MBL patients for the presence of CHD2-mutated cases. This unbiased approach uncovered a total of 5.3% of CLL cases carrying a heterozygous mutation in CHD2, a fairly high percentage when the intrinsic low frequency of mutations in CLL is taken into account.43 Moreover, when corrections by gene size and codon composition are applied, CHD2 is one of the most frequently mutated CLL genes, ranking above other well-defined CLL drivers such as MYD88 and POT1. The link between recurrently mutated CHD2 and B-cell disorders is supported by our finding of 3 mutated cases in a cohort of 43 MBL patients. The nature of most of the CHD2 mutations identified, including truncations, likely elicits protein loss of function, thus suggesting for the first time a tumor suppressor role for CHD2. Notably, CHD1 and CHD5 have been previously identified as tumor suppressors in different malignancies.44,45
Little is known about the biological functions of CHD2. Homozygous Chd2-mutant mice undergo perinatal lethality and heterozygous mice die at early age, mostly because of the development of lymphomas.20,21 CHD2 can act as a positive regulator of myogenic differentiation35 and binds H3K4me3 peptides in vitro, albeit less strongly than CHD1.33 CHD2 has also been described as a regulator with high-specificity binding to active promoters in different cell types including K562 myelogenous leukemia cells and GM12878 lymphoblastoid B cells.46 In this regard, we have observed changes in nuclear distribution of CLL-related CHD2 mutants, with a dramatic effect in the CHD2L1270F variant, which accumulates in the form of large punctated bodies. The severity of the effects observed for CHD2 mutants was variable and related to the specific domains affected, as assessed for CHD2H620L and CHD2F1146L, which exhibit subtle but still apparent nuclear distribution changes. These CHD2 mutations seem to cause defective protein expression, mainly in the case of the CHD2F1146L variant. Notably, the remarkable colocalization between CHD2wt and active chromatin markers was dramatically altered by the DBD mutation L1270F, in which the discrete nuclear bodies seem to be excluded from the loci corresponding to transcriptionally active genes. Co-IP studies confirmed the ability of CHD2 to interact with epigenetic marks for active expression, such as H4Ac and H3K4me3. Because these interactions are conserved in the CHD2H620L and CHD2L1270F mutants, the lack of colocalization of the latter with active chromatin may be explained by a poor or null interaction of the mutated DBD of CHD2L1270F with DNA. Recent studies have confirmed that the C-terminal portion of CHD2 is a functional DBD that displays selectivity for double-stranded DNA and is necessary for the stimulation of the ATPase and chromatin remodeling activities of this protein.47
The aberrant nuclear distribution of CHD2L1270F, together with the loss of association of this mutant with active gene transcription, suggests a functional impact of CHD2 mutations, including truncations, in CLL, likely through transcriptional activation of CLL-relevant genes. In fact, transcriptomic studies and GSEA highlighted significant core enrichment for functional pathways associated with distinct chromatin remodeling processes, from DNA unwinding to the regulation of the acetylation and phosphorylation of histone H3 by the anaphase promoting complex.48 In addition, significant changes are also identified for the clinically relevant PI3K pathway, which lies at the heart of the B-cell receptor signaling pathway and is the subject of a promising CLL targeted therapy based on the PI3K inhibitor idelalisib.38,49
Notably, CHD2 is the most frequently mutated driver gene described in IGHV-mutated CLL cases so far. On the other hand, CHD2-mutated CLL patients do not display any specific clinicobiological signature inside the group of IGHV-mutated cases. Because CLL has become a model disease for the development of personalized medicine, as a result of its clinical and molecular characteristics,1,50 the uncovering of new molecular markers with potentially clinical relevance such as CHD2 sets the basis for the achievement of successful individual CLL therapies. In consequence, a deeper understanding of CHD2 functions in normal and pathological states is warranted for a detailed description of the role of this chromatin remodeler in CLL.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr M. A. Lafarga for advice in the confocal microscopy experiments; D. A. Puente, S. Guijarro, S. Martín, C. Capdevila, M. Osuna, M. Sánchez, and L. Plá for excellent technical assistance; and N. Villahoz and C. Muro for excellent work in the coordination of the CLL Spanish Consortium. The authors also thank all CLL patients who have participated in this study.
This work was supported by the Spanish Ministry of Economy and Competitiveness through the Instituto de Salud Carlos III and Red Temática de Investigación del Cáncer del Instituto de Salud Carlos III. C.L.-O. is an Investigator of the Botín Foundation supported by Banco Santander through its Santander Universities Global Division. E.C. is an Academia Researcher of the Institució Catalana de Recerca i Estudis Avançats de la Generalitat de Catalunya.
Authorship
Contribution: D.R., G.B., and C.L.-O. conceived and designed the experiments and prepared the manuscript; D.R., G.B., V.Q., J.R.A., A.J.R., P.M.Q., and C.R. performed experiments; N.V., A.L.-G., T.B., A.N., J.I.M.-S., and E.C. analyzed data; and C.L.-O. oversaw all of the work.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Carlos López-Otín, Departamento de Bioquímica y Biología Molecular, Universidad de Oviedo, 33006-Oviedo, Spain; e-mail: clo@uniovi.es.
References
Author notes
D.R. and G.B. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal