Key Points
FL-associated STAT6 mutations hyperactivate the IL-4/JAK/STAT6 axis.
Abstract
Follicular lymphoma (FL) is the second most common non-Hodgkin lymphoma in the Western world. FL cell-intrinsic and cell-extrinsic factors influence FL biology and clinical outcome. To further our understanding of the genetic basis of FL, we performed whole-exome sequencing of 23 highly purified FL cases and 1 transformed FL case and expanded findings to a combined total of 114 FLs. We report recurrent mutations in the transcription factor STAT6 in 11% of FLs and identified the STAT6 amino acid residue 419 as a novel STAT6 mutation hotspot (p.419D/G, p.419D/A, and p.419D/H). FL-associated STAT6 mutations were activating, as evidenced by increased transactivation in HEK293T cell–based transfection/luciferase reporter assays, heightened interleukin-4 (IL-4) –induced activation of target genes in stable STAT6 transfected lymphoma cell lines, and elevated baseline expression levels of STAT6 target genes in primary FL B cells harboring mutant STAT6. Mechanistically, FL-associated STAT6 mutations facilitated nuclear residency of STAT6, independent of IL-4–induced STAT6-Y641 phosphorylation. Structural modeling of STAT6 based on the structure of the STAT1-DNA complex revealed that most FL-associated STAT6 mutants locate to the STAT6-DNA interface, potentially facilitating heightened interactions. The genetic and functional data combined strengthen the recognition of the IL-4/JAK/STAT6 axis as a driver of FL pathogenesis.
Introduction
Follicular lymphoma (FL) is the most common indolent B-cell lymphoma, with an incidence of ∼14 000 cases and a prevalence of ∼100 000 cases in the United States.1 FL remains incurable with conventional therapies, and the development of targeted FL-directed therapies is still in the early stages.2-7 FL has a varied clinical course that is influenced by FL tumor cell-intrinsic and cell-extrinsic aberrations.2,5,6,8-10
Cell-intrinsic factors, including genomic and epigenetic deregulations, prominently influence the FL phenotype. At the genomic level, FL is characterized by recurrent acquired structural abnormalities, including acquired genomic copy number aberrations and frequent acquired uniparental disomy/copy neutral loss of heterozygosity.11-15 Furthermore, multiple recurrently mutated genes have recently been shown to underlie the pathogenesis of FL (MLL2, CREBBP, EZH2, ARID1A, HIST1H1, and others).3,16-22
Cell-extrinsic factors, including FL B cell-T cell and B cell-macrophage interactions in the lymph node environment, as well as cytokine-FL cell interactions (collectively referred to as the microenvironment) influence FL biology and outcome.23 One of the ligand-receptor pairs that has an effect on FL B cells and normal B cells is the interleukin-4 (IL-4)/IL-4R pair, which signals through Janus activated kinases (JAKs) and signal transducer of activated T cells 6 (STAT6).24-28 IL-4 levels are elevated in the FL microenvironment because of increased secretion by T follicular helper cells, and a fraction of FL cases harbor nuclear STAT6, indicative of activation.29-31
To further our understanding of the genetic basis of FL and to identify targets for novel therapeutic approaches, we have performed massively parallel exome sequencing of 23 FL cases and one transformed FL (t-FL) case. Through subsequent validation of mutations in 2 expansion cohorts (a combined 114 FL cases), we identified novel STAT6 mutations in 11% of FLs. Functional follow-up studies demonstrated that FL-associated STAT6 mutations are activating and result in an exaggerated transcriptional response to IL-4 in lymphoid tumor cells. Mechanistically, FL-associated STAT6 mutations increased residency of STAT6 in the nucleus involving a novel partially STAT6-Y641 phosphorylation-independent mechanism. In aggregate, these data provide novel insights into the properties of FL-associated STAT6 mutations with broader implications for other lymphoproliferative diseases, including primary mediastinal B-cell lymphoma (PMBCL), diffuse large B-cell lymphoma, and t-FL, carrying such mutations and implications for novel therapy developments targeting the activated IL-4/JAK/STAT6 axis in FL.
Methods
STAT6 expression plasmid cell transfection and luciferase reporter assays
A plasmid containing the STAT6 complementary DNA (cDNA) (NM_003153.4) was purchased from ThermoScientific (Clone ID 5530399) and was used as a template to generate mutant STAT6 cDNAs using the QuikChange Lightning Site-Directed Mutagenesis Kit (Stratagene/Agilent, La Jolla, CA). Full-length wild-type (wt) and mutant influenza hemagglutinin (HA)-tagged STAT6 were constructed by using polymerase chain reaction (PCR) and were cloned into the PacI/NotI sites of the lentiviral vector FG9 (a gift from Dr Colin Duckett, University of Michigan32 ). The luciferase reporter plasmid pGL3-Luc encoding one wt or mutated copy of a STAT6-responsive element (a 1.095-kb genomic fragment of the eotaxin-3 gene containing a single active STAT6 binding site [TTCTCTGGAA] or a mutated binding site [AGCTCTGGAA]) upstream of the firefly luciferase coding gene was a generous gift from Dr Zhengfan Zhang (Peking University, China).33 Details of all luciferase constructs used in this study are provided in supplemental Methods available online at the Blood Web site.
HEK293T cells were transfected in duplicate with 0.1 µg of STAT6 luciferase reporter plasmid, together with 0.9 µg of plasmids encoding either wt or mutant forms of STAT6 (all HA-tagged) and Renilla luciferase (R-Luc) using polyethylenimine (Polyscience Inc. #23966). Twenty-four hours later, IL-4 (Life Technologies #11846-5) at 10 ng/mL was added to parallel cultures for 6 hours, cell lysates were prepared, and luciferase activities were measured with a Dual-Luciferase assay kit (Promega, Madison, WI). Anti-STAT6 antibody was purchased from Santa Cruz Biotechnology (Dallas, TX; #SC-621), anti-phospho-STAT6-Y641 antibody was from Cell Signaling Technologies (Danvers, MA; #9361S), anti-HA antibody was from Roche (Basel, Switzerland; clone 3F10; #1867423001), and anti-tubulin and anti-lamin B1 antibodies were from Santa Cruz Biotechnology (#SC-32293 and #SC-377000, respectively) (supplemental Methods).
Results
Massively parallel sequencing of the coding genome of FL samples
To further our understanding of the genetic basis of FL, we used solution exon capture of sheared and processed genomic DNA isolated from flow-sorted immunoglobulin light chain–restricted lymphomatous B cells and paired CD3+ T cells isolated from 23 patients with FL and 1 patient with diffuse large B-cell lymphoma transformed from prior FL (t-FL) followed by paired-end massively parallel sequencing. DNA purity was estimated at 77% to 100% with a mean of 91% (supplemental Table 1). The sequence data were characterized by a range of de-duplicated mapped reads per DNA sample of 28 554 787 to 68 864 168 (mean, 43 871 048) and a depth of coverage range of 23 to 81 (mean of 45), mean number of de-duplicated reads per nucleotides in the exome. Data from the initial analysis of 11 FL and 1 t-FL patient have been published.21
Identification of recurrent somatic mutations in STAT6 in FL
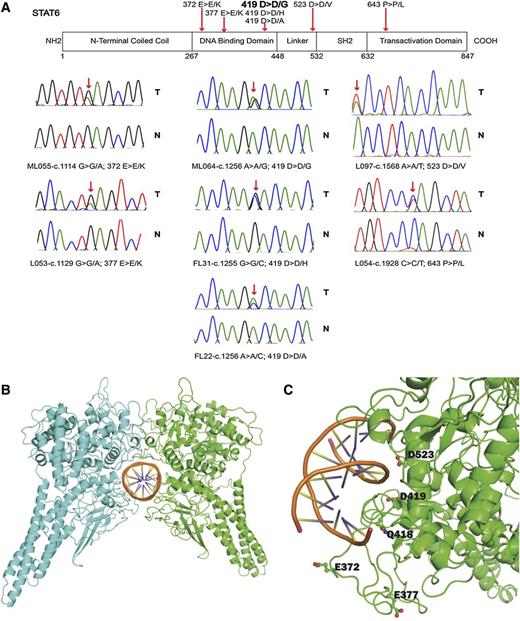
Within the discovery group of 23 FL cases and 1 t-FL case, we identified two STAT6 mutations, one of which occurred in patient ML55 (t-FL) as previously described.21 Resequencing of all STAT6 coding exons in a combined total of 113 FL and 1 t-FL patients identified a total of 11% (12 of 114) of patients with nonsynonymous STAT6 mutations, one of whom (L54) contained 2 mutations (Figure 1A and Table 1). All STAT6 mutations were heterozygous. The majority of the STAT6 mutations targeted the STAT6 DNA binding domain, whereas 1 mutation (p.523D>V) was located in the linker domain and another (p.643P>L) targeted a residue of 2 amino acids C-terminal to the critical JAK phosphorylation site in STAT6 (Y641). Within the spectrum of STAT6 mutations, we identified a novel STAT6 mutation hotspot in amino acid residue 419, which was mutated from aspartic acid (D) to either glycine (G), or histidine (H), or alanine (A). The mutation STAT6 p.419D>D/G was the most common recurrent STAT6 mutation identified.
Identification of novel mutations in STAT6 in FL. (A) Schema of the STAT6 protein domain structure. The approximate location of somatic mutations identified in STAT6 in FL is indicated. Sanger sequence chromatograms showing results for FL B-cell– and paired CD3+ T-cell–derived DNA. (B-C) Homology model of the STAT6-DNA complex. (B) STAT6 dimer with individual monomers in green and cyan. (C) STAT6 residues undergoing mutation are close to the DNA binding interface and are displayed in ball-and-stick diagrams. The graphic was generated with the PyMOL program.
Identification of novel mutations in STAT6 in FL. (A) Schema of the STAT6 protein domain structure. The approximate location of somatic mutations identified in STAT6 in FL is indicated. Sanger sequence chromatograms showing results for FL B-cell– and paired CD3+ T-cell–derived DNA. (B-C) Homology model of the STAT6-DNA complex. (B) STAT6 dimer with individual monomers in green and cyan. (C) STAT6 residues undergoing mutation are close to the DNA binding interface and are displayed in ball-and-stick diagrams. The graphic was generated with the PyMOL program.
Details of gene mutations in FL patients carrying STAT6 mutations
| FL Patient No. . | Disease status . | STAT6 cDNA . | STAT6 . | CREBBP . | MLL2 . | TNFRSF14 . | EZH2 . | MEF2B . | HIST1H1 B-E . | ARID1A . | TP53 . | OCT2 . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ML055 | Relapsed | c.1114 G>G/A | p.372 E>E/K | p.1472 W>W/S p.1103 R>R/X | p.3789fs | p.112 S>F | p.641 Y>Y/N | wt | wt | wt | wt | wt |
| L053 | Relapsed | c.1129 G>G/A | p.377 E>E/K | p.1446 R>R/C | p.3681 Q>Q/X; p.Leu 925fs | wt | wt | wt | HIST1H1-D p.102A>A/V | wt | wt | wt |
| FL25 | Unknown | c.1129 G>G/A | p.377 E>E/K | p.200 Q>X | wt | p.12 W>W/X p.69 E>E/X | wt | wt | wt | wt | wt | wt |
| L026 | Untreated | c.1255 G>G/C | p.419 D>D/H | Exon 28 splice donor mutation | p.3988 Q>Q/X | wt | wt | wt | wt | wt | wt | wt |
| FL31 | Unknown | c.1255 G>G/C | p.419 D>D/H | p.1543 D>D/V | p.3395 Q>Q/X | p.1 M>I | p.641 Y>Y/F | wt | wt | wt | wt | wt |
| FL38 | Unknown | c.1255 G>G/C | p.419 D>D/H | p.1502 W>R | wt | p.158 Q>X | wt | wt | wt | wt | wt | wt |
| FL22 | Unknown | c.1256 A>A/C | p.419 D>D/A | p.1499 L>L/P | Exon 15 splice donor mutation | wt | wt | wt | wt | wt | wt | wt |
| ML064 | Untreated | c.1256 A>A/G | p.419 D>D/G | p.1421 C>C/F; p.482 Q>Q/X | wt | wt | wt | wt | HIST1H1-D p.84G>D | wt | wt | wt |
| L084 | Relapsed | c.1256 A>A/G | p.419 D>D/G | p.1446 R>L | wt | wt | wt | wt | wt | wt | p.262 G>V | wt |
| L046 | Untreated | c.1256 A>A/G | p.419 D>D/G | p.1446 R>C | wt | p.3fs | wt | wt | HIST1H1-D p.82K>K/D | wt | wt | p.223T>T/S |
| L054 | Relapsed | c.1256 A>A/G; c.1928 C>C/T | p.419 D>D/G; p.643 P>P/L | p.S1680* | p.2970 S>S/N | p.96 C>X | p.641 Y>Y/N | wt | wt | wt | wt | wt |
| L097 | Untreated | c.1568 A>A/T | p.523 D>D/V | p.1484 F>F/S | p.2685 R>R/X | wt | wt | wt | wt | wt | wt | wt |
| FL Patient No. . | Disease status . | STAT6 cDNA . | STAT6 . | CREBBP . | MLL2 . | TNFRSF14 . | EZH2 . | MEF2B . | HIST1H1 B-E . | ARID1A . | TP53 . | OCT2 . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ML055 | Relapsed | c.1114 G>G/A | p.372 E>E/K | p.1472 W>W/S p.1103 R>R/X | p.3789fs | p.112 S>F | p.641 Y>Y/N | wt | wt | wt | wt | wt |
| L053 | Relapsed | c.1129 G>G/A | p.377 E>E/K | p.1446 R>R/C | p.3681 Q>Q/X; p.Leu 925fs | wt | wt | wt | HIST1H1-D p.102A>A/V | wt | wt | wt |
| FL25 | Unknown | c.1129 G>G/A | p.377 E>E/K | p.200 Q>X | wt | p.12 W>W/X p.69 E>E/X | wt | wt | wt | wt | wt | wt |
| L026 | Untreated | c.1255 G>G/C | p.419 D>D/H | Exon 28 splice donor mutation | p.3988 Q>Q/X | wt | wt | wt | wt | wt | wt | wt |
| FL31 | Unknown | c.1255 G>G/C | p.419 D>D/H | p.1543 D>D/V | p.3395 Q>Q/X | p.1 M>I | p.641 Y>Y/F | wt | wt | wt | wt | wt |
| FL38 | Unknown | c.1255 G>G/C | p.419 D>D/H | p.1502 W>R | wt | p.158 Q>X | wt | wt | wt | wt | wt | wt |
| FL22 | Unknown | c.1256 A>A/C | p.419 D>D/A | p.1499 L>L/P | Exon 15 splice donor mutation | wt | wt | wt | wt | wt | wt | wt |
| ML064 | Untreated | c.1256 A>A/G | p.419 D>D/G | p.1421 C>C/F; p.482 Q>Q/X | wt | wt | wt | wt | HIST1H1-D p.84G>D | wt | wt | wt |
| L084 | Relapsed | c.1256 A>A/G | p.419 D>D/G | p.1446 R>L | wt | wt | wt | wt | wt | wt | p.262 G>V | wt |
| L046 | Untreated | c.1256 A>A/G | p.419 D>D/G | p.1446 R>C | wt | p.3fs | wt | wt | HIST1H1-D p.82K>K/D | wt | wt | p.223T>T/S |
| L054 | Relapsed | c.1256 A>A/G; c.1928 C>C/T | p.419 D>D/G; p.643 P>P/L | p.S1680* | p.2970 S>S/N | p.96 C>X | p.641 Y>Y/N | wt | wt | wt | wt | wt |
| L097 | Untreated | c.1568 A>A/T | p.523 D>D/V | p.1484 F>F/S | p.2685 R>R/X | wt | wt | wt | wt | wt | wt | wt |
ML055 mutation context: c.11358-11366dupGGTCCAGC; L026 mutation context: AACCACTGAGctacagaccct; FL22 mutation context: AGTGCAAGTGttaaggagact; L054 mutation context: c.5039_5041delCCT. CREBBP cDNA: NM_004380.2; MLL2 cDNA: NM_003482.3; TNFRSF14 cDNA: NM_003820.2; MEF2B cDNA: NM_001145785.1.
fs, frameshift.
A subsequent review of common FL-associated mutations in FL patients carrying STAT6 mutations demonstrated that all such patients carried CREBBP mutations and none carried mutations in ARID1A or MEF2B (Table 1).
FL-associated STAT6 mutations are located on the surface of the STAT6 protein in close contact with DNA
We generated 3-D modeling data of STAT6 bound to the STAT6 binding site TTCTCTGGAA. In the absence of an experimentally determined structure of STAT6, a homology model based on STAT1 was developed.34 STAT6 residues E372, E377, Q418, D419, and D523, which are part of the STAT6 DNA binding domain, were located along the surface of the protein at the DNA-protein interface (Figure 1B-C). Residue D419 locates along the major groove, whereas E372, E377, and D523 locate along the minor groove. The proximity of the residues to DNA suggests that mutations of these residues might have a direct influence on the DNA binding of STAT6.
Intrinsic activation of FL-associated STAT6 mutants as detected through HEK293T cell luciferase assays
To derive initial insights into the properties of FL-associated STAT6 mutant proteins, we used HEK293T cells because they are widely used for promoter-driven luciferase assays, they lack endogenous STAT6, and they have a functional IL-4/JAK pathway.33 We cotransfected empty FG9 vector, FG9-HA-STAT6-wt, or FG9-HA-STAT6 mutants together with either a wt or mutated (TTCTCTGGAA changed to AGCTCTGGAA) STAT6 response plasmid33 into HEK293T cells and, after 24 hours, we added IL-4 for 6 hours to parallel cultures. Subsequently, we measured luciferase activity in cell lysates that were prepared in the presence of sodium orthovanadate.
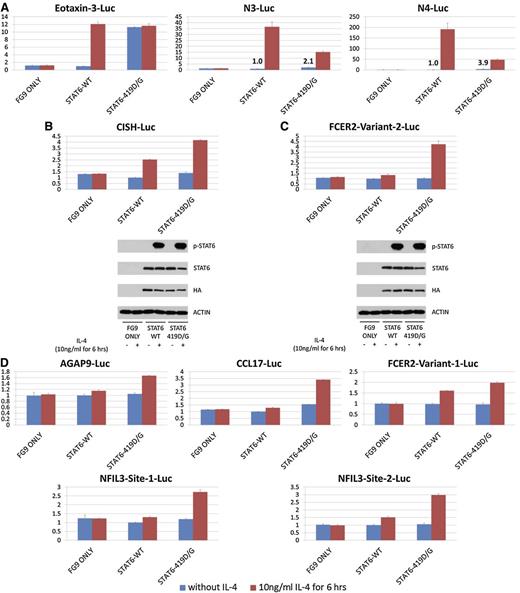
IL-4 addition resulted in a strong (∼11-fold) induction by STAT6 wt of the eotaxin-3 gene luciferase response plasmid, which was absent when a mutated STAT6 responsive element containing plasmid was used (Figure 2A and supplemental Figure 1). Most STAT6 mutants were also IL-4 responsive, but results were most notable for a substantial intrinsic activation of the STAT6 mutants that was independent of IL-4 (n = 10 separate experiments). We measured an intrinsic STAT6 activation across the various mutants and across the various experiments ranging from approximately threefold to 16-fold. Immunoblotting for STAT6-Y641 did not identify baseline phosphorylation of any of the STAT6 proteins, implying that the STAT6 mutations activate STAT6 proteins through a novel mechanism (Figure 2C).
FL-associated STAT6 mutations are intrinsically activating and partially STAT6-Y641 independent. Results of STAT6 luciferase assays in HEK293T cells and corresponding immunoblots. (A) Luciferase assay results in HEK293T cells indexed to the results for unstimulated STAT6 wt measurements; IL-4 stimulation is indicated. (B) Immunoblot results for the experiment presented in (A) using antibodies against STAT6, p-STAT6-Y641, and actin. (C) Luciferase assay results in HEK293T cells indexed to the results for unstimulated STAT6 wt measurements; IL-4 stimulation is indicated. (D) Immunoblot results for the experiment presented in (C) using antibodies against STAT6, p-STAT6 Y641, and actin.
FL-associated STAT6 mutations are intrinsically activating and partially STAT6-Y641 independent. Results of STAT6 luciferase assays in HEK293T cells and corresponding immunoblots. (A) Luciferase assay results in HEK293T cells indexed to the results for unstimulated STAT6 wt measurements; IL-4 stimulation is indicated. (B) Immunoblot results for the experiment presented in (A) using antibodies against STAT6, p-STAT6-Y641, and actin. (C) Luciferase assay results in HEK293T cells indexed to the results for unstimulated STAT6 wt measurements; IL-4 stimulation is indicated. (D) Immunoblot results for the experiment presented in (C) using antibodies against STAT6, p-STAT6 Y641, and actin.
We cotransfected STAT6 wt and STAT6 419G in various ratios into HEK293T cells and measured the transactivation output by using the eotaxin-3 luciferase activation assay (n = 3). An equimolar ratio of STAT6 wt and STAT6 419G at baseline (no IL-4 stimulation) produced less than half the transactivation output of the STAT6 419G-alone transfection (supplemental Figure 2), suggesting that the presence of STAT6 wt proteins partially interfered with the STAT6 mutant protein’s ability to transactivate.
We detected equal IL-4–induced STAT6-Y641 phosphorylation in STAT6 wt and STAT6 mutant proteins at the end of the 6-hour stimulation, and expression levels of STAT6 were equal (Figure 2B,D). However, when performing kinetic analyses of IL-4–induced STAT6-Y641 phosphorylation (15 minutes to 8 hours), we noticed substantially decreased STAT6-Y641 phosphorylation of the STAT6 372K and 419G mutants when compared with STAT6 wt in the first few hours after IL-4 stimulation, which equalized at later time points (supplemental Figure 3).
STAT6 double mutants harboring STAT6-Y641F remain partially activating
We created artificial STAT6 double mutants each containing 1 of 3 STAT6 mutations: p.372E>K, p.377E>K, or p.419D>G as well as the mutation p.641Y>F, and tested the activity of these mutants in the eotaxin-3 gene luciferase activation assay (n = 3). In response to IL-4, the STAT6-Y641F mutant was inactive in the assay (Figure 2C). In contrast, the effect of the Y641F substitution in the FL-associated STAT6 mutants was complex: it reduced the IL-4–independent intrinsic activation potential of these mutants compared with the single mutants but still demonstrated a substantially higher IL-4–independent intrinsic activation than the STAT6 wt protein, and it abolished the IL-4–induced activation of these mutants beyond their intrinsic activation potential (Figure 2C).
Results of analyses of the STAT6 419G mutant on an extended panel of luciferase response plasmids in HEK293T cell luciferase assays
Published luciferase reporter assay results using the STAT6 D419H mutation demonstrated reduced maximal IL-4–induced activation compared with STAT6 wt when assayed on response plasmids carrying synthetic response elements harboring 3 STAT6 binding sites.35 We assayed the STAT6 D419G mutant using these plasmids and confirmed this observation. In addition, we detected IL-4–independent increased baseline activation of STAT6 D419G (Figure 3A).
Results of STAT6 luciferase assays in HEK293T cells across a panel of luciferase response plasmids. (A) Results for an eotaxin-3 luciferase (Luc) plasmid and two synthetic STAT6 luciferase response plasmids described by Ritz et al,35 each carrying 3 STAT6 sites (supplemental Methods); N3: TTCNNNGAA; N4: TTCNNNNGAA. Numbers indicate fold-activation over the unstimulated STAT6 wt signal. (B-C) Results for luciferase assays using naturally occurring genomic DNA stretches carrying STAT6 site(s) derived from CISH and FCER2 and corresponding immunoblot results. (D) Results for luciferase assays for 5 naturally occurring genomic DNA stretches carrying single STAT6 sites in AGAP9, CCL17, FCER2, and two NFIL3-derived sites.
Results of STAT6 luciferase assays in HEK293T cells across a panel of luciferase response plasmids. (A) Results for an eotaxin-3 luciferase (Luc) plasmid and two synthetic STAT6 luciferase response plasmids described by Ritz et al,35 each carrying 3 STAT6 sites (supplemental Methods); N3: TTCNNNGAA; N4: TTCNNNNGAA. Numbers indicate fold-activation over the unstimulated STAT6 wt signal. (B-C) Results for luciferase assays using naturally occurring genomic DNA stretches carrying STAT6 site(s) derived from CISH and FCER2 and corresponding immunoblot results. (D) Results for luciferase assays for 5 naturally occurring genomic DNA stretches carrying single STAT6 sites in AGAP9, CCL17, FCER2, and two NFIL3-derived sites.
Next, multiple luciferase response plasmids were constructed that carried naturally occurring genomic DNA sequences harboring STAT6 sites (supplemental Methods) derived from either promoters or sites downstream from transcription start sites from the STAT6-responsive genes CISH, FCER2, CCL17, NFIL3, and AGAP9. In all tested conditions, the STAT6 419G mutant, either at baseline or more conspicuous in response to IL-4, was more active than STAT6 wt (Figure 3B-D).
We performed electrophoretic mobility shift assays using lysates from HEK293T cells transfected with empty FG9 vector, FG9-HA-STAT6-wt, or FG9-HA-STAT6 419G and oligonucleotides carrying the single STAT6 sites present in the CCL17 or FCER2 constructs. Results demonstrated increased binding of STAT6-419G as opposed to STAT6 wt (supplemental Figure 4).
Results of expression array analysis of stable lymphoma cell lines transduced with STAT6 wt or STAT6 mutants
To analyze the properties of FL-associated STAT6 mutants (372K, 377K, and 419G) in lymphoid cell lines that express endogenous STAT6, we generated stable lymphoma cell lines (OCI-LY1, OCI-LY7, and OCI-LY8). The resulting stable lines expressed appreciable quantities of exogenous STAT6 (supplemental Figure 5). Note that OCI-LY1 contained a STAT6 variant (p.375G>R) that was not detected in any of the FL patients. We assayed the activity of this variant across 6 reporter constructs in HEK293T cells. In some but not all assays, a very modest increase in either baseline or stimulated activity compared with STAT6 wt was noted (supplemental Figure 6).
Next, we assessed the degree and kinetics of STAT6-Y641 phosphorylation after stimulation with IL-4. Lysates prepared in 100 mM NaCl demonstrated reduced Y641 phosphorylation of the STAT6 mutants compared with wt STAT6 (supplemental Figure 7, upper panel); this difference was not evident in lysates prepared in 400 mM NaCl (supplemental Figure 7, lower panel), supporting the conclusion that some of the STAT6 mutants had altered physical properties and were engaged in salt-sensitive molecular interactions.
We performed an expression array experiment on RNA isolated from all lines at baseline (no exogenous IL-4 addition and IL-4 concentration in the serum-supplemented media below 2 pg/mL as measured by enzyme-linked immunosorbent assay) as well as from a subset of the lines harboring empty vector, STAT6 wt, or STAT6 419G after stimulation for 6 hours with 10 ng/mL of IL-4. Resulting expression values were analyzed as described, and probes were further categorized according to the presence of experimentally verified IL-4–responsive STAT6 binding sites (supplemental Methods).
We compared results for baseline expression data for all 3 STAT6 mutants in all 3 lines combined (9 expression sets) with the data for lines harboring vector alone; in this analysis, we did not detect evidence for IL-4–independent STAT6 mutant-dependent transcriptome changes in genes containing STAT6 binding sites.
Next, we compared the transcriptome changes after IL-4 treatment of cell lines expressing HA-STAT6 wt vs HA-STAT6-419G. IL-4 stimulation of HA-STAT6 wt transfectants resulted in changes in gene expression in 451 protein coding genes or microRNAs, 17 of which contained experimentally verified IL-4–responsive STAT6 binding sites (supplemental Table 2). IL-4 stimulation of HA-STAT6 419G transfectants resulted in expression changes in 450 protein coding genes or microRNAs, 23 of which contained experimentally verified IL-4–responsive STAT6 binding sites (supplemental Table 3). A direct comparison of both transcriptome changes identified 126 shared deregulated genes; 7 of these shared genes contained experimentally verified IL-4–responsive STAT6 binding sites.
Various gene set enrichment analyses (GSEAs) of the expression array data sets were performed.36 GSEA data sets from unstimulated cells (eg, STAT6 419G vs STAT6 wt or, alternatively, vector alone) did not identify enriched sets, consistent with the analyses detailed above. GSEA performed on data sets from IL-4–stimulated experiments detected a substantial number of expected significantly enriched sets in both STAT6 wt and STAT6 419G stable lines, many of which were shared (supplemental Table 4). Overall, a larger number of sets were uniquely enriched in the IL-4–treated STAT6 419G stable lines than in the wt STAT6 lines, which we suspect was a result of augmented IL-4 signaling in the STAT6 419G stable lines. Of interest, shared by both sets was a prominent enrichment for HIF1 and HIF2 targets, pointing toward a heretofore unidentified intersection of the IL-4/JAK/STAT6 and HIF pathways in lymphoma that deserves further study.
STAT6 mutants confer enhanced responsiveness to IL-4 in lymphoma cell lines
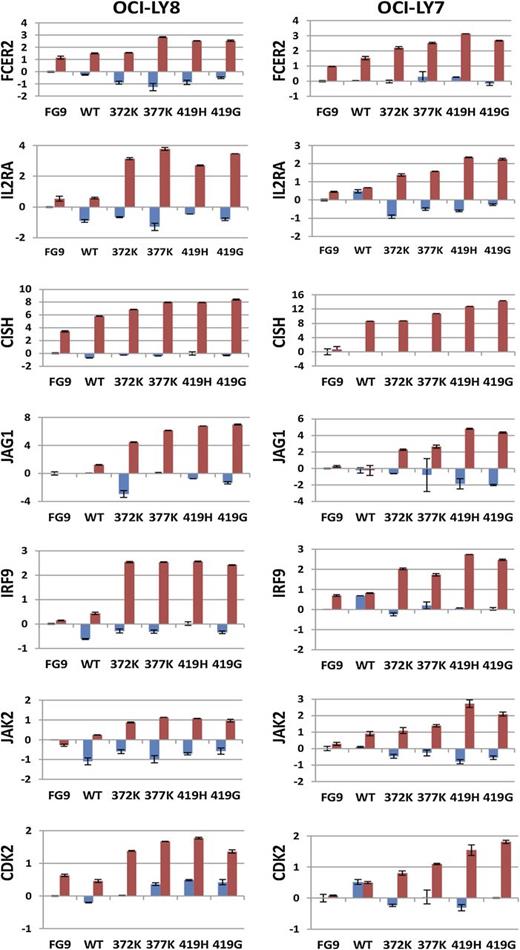
IL-4 treatment of the cell lines stably transfected with STAT6 419G resulted in changes in expression of multiple genes with interesting biological properties: IL2RA, JAG1 (a NOTCH ligand), IRF9 (a STAT5 binding protein), JAK2, and CDK2, as well as the known STAT6 response genes CISH and FCER2. cDNA was made from total RNA from OCI-LY7 and OCI-LY8 lines (containing FG9, STAT6 wt, or STAT6 372K, 377K, 419G, or 419H) that were either left untreated or were stimulated with IL-4. We measured the expression of these 7 genes by quantitative PCR (qPCR) (Figure 4). With some heterogeneity noted, most of these genes demonstrated elevated or substantially elevated expression in IL-4–treated mutant STAT6 cell lines when compared with the IL-4–treated STAT6 wt transfected cell lines or the vector control lines, providing evidence that FL-associated STAT6 mutants in this experimental setting conferred heightened responsiveness to IL-4 (please note that eotaxin-3 was not expressed in either of these cell lines, and baseline CISH expression was below detection in OCI-LY7). In agreement with the above-mentioned array-based measurements, the baseline expression of these target genes was not substantially altered by STAT6 mutants.
Heightened expression of 7 genes in response to IL-4 in lymphoid cell lines overexpressing STAT6 mutants. The stable cell lines OCI-LY7 and OCI-LY8, expressing various STAT6 constructs as indicated, were left unstimulated (blue) or stimulated with IL-4 (red), and the expression of 7 genes was measured by qPCR. Data were normalized to the expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and to the measurements for the unstimulated FG9 (vector only) cells. Displayed are δ Ct values. The mean is indicated.
Heightened expression of 7 genes in response to IL-4 in lymphoid cell lines overexpressing STAT6 mutants. The stable cell lines OCI-LY7 and OCI-LY8, expressing various STAT6 constructs as indicated, were left unstimulated (blue) or stimulated with IL-4 (red), and the expression of 7 genes was measured by qPCR. Data were normalized to the expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and to the measurements for the unstimulated FG9 (vector only) cells. Displayed are δ Ct values. The mean is indicated.
We also measured the expression of IRF9, CISH, JAG1, FCER2, JAK2, CDK2, and CCL11 (eotaxin-3) from their endogenous gene loci in HEK293T cells before and after stimulation with IL-4. However, with the exception of minor expression increases following IL-4 in CISH, none of these genes were inducible in these nonlymphoid cells.
The direct STAT6 target genes CISH, FCER2, NFIL3, and CCL17 are overexpressed in STAT6 mutant primary FL B cells compared with STAT6 wt FL B cells
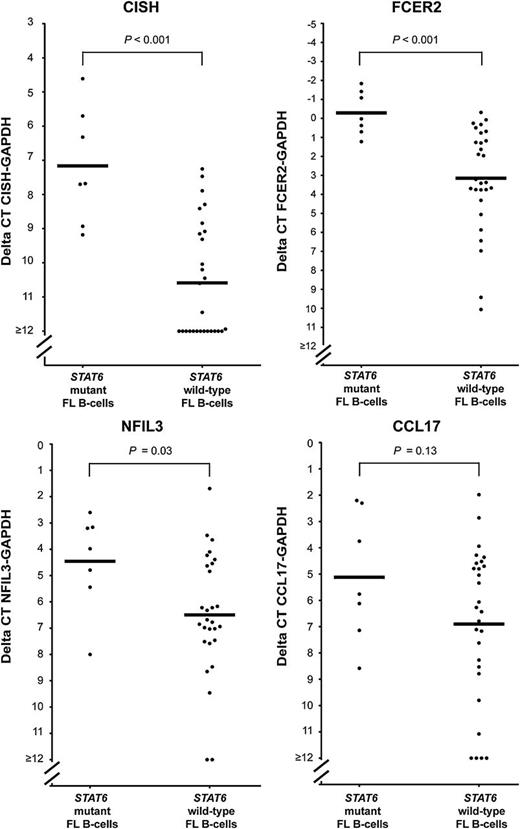
By using random-primed cDNA made from RNA isolated from flow-sorted FL B cells harboring STAT6 mutants or STAT6 wt, we measured the baseline (no exogenous IL-4 stimulation) expression of CISH, FCER2, NFIL3, and CCL17 by qPCR. As can be seen in Figure 5, expression of CISH, FCER2, and NFIL3 (displayed as δ Ct values on a log2 scale) was significantly and markedly higher in STAT6 mutant FL B cells when compared with STAT6 wt B cells, and CCL17 levels were higher.
Elevated baseline expression of CISH, FCER2, NFIL3, and CCL17 in primary FL B cells carrying mutated STAT6. The expression of CISH, FCER2, NFIL3, and CCL17 was measured by qPCR in cDNA made from total RNA isolated from flow-sorted lymphoma B cells. Displayed are δ Ct values. The mean is indicated.
Elevated baseline expression of CISH, FCER2, NFIL3, and CCL17 in primary FL B cells carrying mutated STAT6. The expression of CISH, FCER2, NFIL3, and CCL17 was measured by qPCR in cDNA made from total RNA isolated from flow-sorted lymphoma B cells. Displayed are δ Ct values. The mean is indicated.
Next, we purified B cells from cryopreserved FL lymph nodes containing wt (n = 6) or mutant (n = 4; only 4 patients had sufficient material for analyses) STAT6 using column- and bead-based negative selection. Cells were rested for 2 hours in serum-supplemented medium, and aliquots were subsequently stimulated with 10 ng/mL IL-4 for 4 hours. Total RNA was extracted, cDNA was made, and the normalized expression of BCL2L1, FCER2, CCL17, CISH, and NFIL3 was measured by qPCR. The average IL-4 inducibility of these genes was largely of similar magnitude in both STAT6 mutant and wt cases with mutants demonstrating elevated baseline expression (supplemental Figure 8).
FL-associated mutant STAT6 proteins preferentially localize to the nucleus
Given the central physiological role of cytoplasmic/nuclear shuttling of STAT6 after IL-4 stimulation,37 we measured the subcellular localization of STAT6 in transfected HEK293T cells and in stably transfected OCI-LY7 and OCI-LY8 cell lines by using biochemical subcellular fractionation before and after IL-4 treatment. Before IL-4 stimulation, we detected a fraction of STAT6 mutant proteins in the nucleus as opposed to the STAT6 wt protein, which was largely cytoplasmic (Figure 6A). After IL-4 stimulation, most of the Y641-phosphorylated STAT6 proteins were located in the nucleus, resulting in a modest increase in the mutant protein fraction localizing to the nucleus. In addition, a substantial fraction of the STAT6 419G/641F double mutant (but not the STAT6 641F single mutant) localized to the nucleus, providing evidence for a novel mechanism of nuclear localization of STAT6 419G proteins. Similar results were obtained when we analyzed HEK293T cells transfected with STAT6 wt or STAT6 mutant constructs.
A greater propensity for STAT6 mutant proteins vs STAT6 wt proteins to reside in the nucleus before and after IL-4 stimulation. (A-C) Subcellular fractionation. Stably transfected lymphoma cell lines OCI-LY7 and OCI-LY8 and transiently transfected HEK293T cells were fractionated by using hypotonic cell lysis (cytoplasmic fraction) followed by high salt extractions (nuclear fraction), and protein was prepared for immunoblotting with antibodies to HA, p-STAT6-Y641, laminB1, and tubulin. IL-4 stimulation and STAT6 mutant and double mutant status is indicated at the top. (D) Immunofluorescence. Results of confocal immunofluorescence detection of HA-tagged STAT6 wt and STAT6 419G in stably transfected lymphoma cells and transiently transfected HEK293T cells. Red, anti-HA; blue, 4,6 diamidino-2-phenylindole staining. IL-4 stimulation is indicated.
A greater propensity for STAT6 mutant proteins vs STAT6 wt proteins to reside in the nucleus before and after IL-4 stimulation. (A-C) Subcellular fractionation. Stably transfected lymphoma cell lines OCI-LY7 and OCI-LY8 and transiently transfected HEK293T cells were fractionated by using hypotonic cell lysis (cytoplasmic fraction) followed by high salt extractions (nuclear fraction), and protein was prepared for immunoblotting with antibodies to HA, p-STAT6-Y641, laminB1, and tubulin. IL-4 stimulation and STAT6 mutant and double mutant status is indicated at the top. (D) Immunofluorescence. Results of confocal immunofluorescence detection of HA-tagged STAT6 wt and STAT6 419G in stably transfected lymphoma cells and transiently transfected HEK293T cells. Red, anti-HA; blue, 4,6 diamidino-2-phenylindole staining. IL-4 stimulation is indicated.
We also used immunofluorescence and confocal microscopy to locate HA-tagged STAT6 proteins in transfected HEK293T cells as well as in the stably transfected lymphoma cell lines by using an anti-HA antibody, despite potential limits to detect nuclear, dimerized, and DNA-bound STAT6 quantitatively (Figure 6D). Empty vector (FG9) transfected cells showed faint background staining. Cells transfected with HA-STAT6 wt or HA-STAT6-419G demonstrated predominantly cytoplasmic staining with occasional cells also showing nuclear signals. Upon IL-4 stimulation, the nuclear staining compared with the cytoplasmatic staining was much stronger in the STAT6 419G mutant as opposed to the STAT6 wt transfectants.
Lentiviral-mediated STAT6 knockdown in lymphoma cell lines induces apoptosis
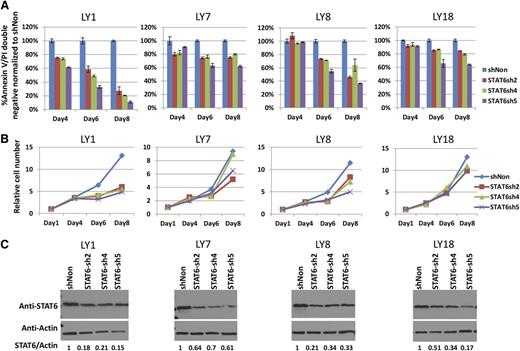
Potent short hairpin RNA targeting the STAT6 open reading frame or untranslated region were identified (supplemental Figure 9). Next, lentiviral transduction of these short hairpin RNAs into the cell lines OCI-LY1, OCI-LY7, OCI-LY8, and OCI-LY18 was performed (day 1). The viability of the infected bulk cell population was measured by annexinV/propidium iodide staining on days 4, 6, and 8, and corresponding cell numbers were determined. As can be seen in Figure 7, all cell lines were dependent on STAT6 to varying degrees for survival. Given the lack of exogenous IL-4 supplementation and the measured IL-4 levels in the medium below the detection limit of 2 pg/mL, these data imply a tonic survival function of STAT6 in these cell lines reminiscent of what has been described in Hodgkin lymphoma cell lines.38
Targeting STAT6 by using short hairpin RNAs (shRNAs) induces apoptosis in lymphoma cell lines. (A) The lymphoma cell lines OCI-LY1, OCI-LY7, OCI-LY8, and OCI-LY18 were transduced with lentiviruses carrying 3 distinct STAT6-targeted shRNAs (day 1). On days 4, 6, and 8 after infection, the fraction of annexinV/propidium iodide double-negative cell populations was measured and data were plotted normalized to the double-negative fraction in the sh-nontargeted (shNon) control cell population. (B) Cells were diluted 1:1 in all cultures every 2 days starting on day 4. The relative cell number as measured through automated cell counters corrected for the dilutions is plotted. (C) Immunoblot and densitometry results for STAT6 knockdown in bulk populations on day 4 after infection.
Targeting STAT6 by using short hairpin RNAs (shRNAs) induces apoptosis in lymphoma cell lines. (A) The lymphoma cell lines OCI-LY1, OCI-LY7, OCI-LY8, and OCI-LY18 were transduced with lentiviruses carrying 3 distinct STAT6-targeted shRNAs (day 1). On days 4, 6, and 8 after infection, the fraction of annexinV/propidium iodide double-negative cell populations was measured and data were plotted normalized to the double-negative fraction in the sh-nontargeted (shNon) control cell population. (B) Cells were diluted 1:1 in all cultures every 2 days starting on day 4. The relative cell number as measured through automated cell counters corrected for the dilutions is plotted. (C) Immunoblot and densitometry results for STAT6 knockdown in bulk populations on day 4 after infection.
Discussion
We present the discovery and initial functional characterization of heterozygous mutations in STAT6 in 11% of FL. FL-associated STAT6 mutations predominantly targeted the DNA-binding domain in a novel STAT6 mutation hotspot in amino acid residue 419, which was mutated from aspartic acid (D) to glycine (G), histidine (H), or alanine (A). STAT6 mutations have recently been reported by Okuson and colleagues in 12% of FL patients and by Pasqualucci and colleagues in 23% of t-FL patients; combined, these findings validate STAT6 as a novel frequently mutated gene in FL/t-FL.20,22
The functional data presented in this study support the conclusion that FL-associated STAT6 mutations are activating, and they complement existing data on the importance of an activated IL4/JAK/STAT6 axis in FL and other non-Hodgkin lymphoma.38,39 This conclusion is supported by the following experimental findings: (1) substantial intrinsic activation of the STAT6 mutants when assayed on a genomic fragment containing the STAT6 consensus binding site TTCTCTGGAA, (2) enhanced IL-4–induced activation of various luciferase constructs derived from known STAT6 responsive genes, (3) stronger induction of multiple IL-4 target genes in lymphoma cells stably transfected with STAT6 mutants vs wt STAT6, (4) substantially elevated baseline expression levels of STAT6 target genes in unstimulated sorted primary FL B cells carrying mutated STAT6 as opposed to cells carrying STAT6 wt, and (5) augmented baseline and stimulated nuclear localization of mutant STAT6 proteins.
Previous work demonstrated elevated IL-4 levels in the FL tumor microenvironment as well as nuclear STAT6 staining in a fraction of FL B cells, indicating that an activated IL-4/JAK/STAT6 axis characterizes FL biology.29 Other lines of evidence supporting a broader role for various augmented JAK/STAT pathways in lymphoproliferative diseases include recurrent mutations in SOCS1 in FL,22,40 genomic gains of JAK2 in both PMBCL and Hodgkin lymphoma,41 mutations in PTPN1 in PMBCL and Hodgkin lymphoma,42 activated STAT3 in CLL,43 and nuclear STAT6 in PMBCL.44 In contrast, published data on the STAT6 mutations occurring in PMBCL were interpreted to indicate hypomorphic functions for the STAT6 mutant proteins.35
The discovery of the activating properties of FL-associated STAT6 mutations originated with results of a study that used a published eotaxin-3 response plasmid (see Figure 2).33 When we subsequently assayed the STAT6 mutants on two synthetic response plasmids generated by Ritz and colleagues35 each containing 3 STAT6 sites, the STAT6 wt protein induced a stronger IL-4–dependent response as compared with the STAT6 419G mutant. However, we noticed an IL-4–independent baseline activation of the STAT6 mutants. Luciferase reporter assays on an extended panel of naturally occurring genomic STAT6 sites derived from FCER2, CISH, NFIL3, CCL17, and AGAP9 demonstrated heightened responsiveness of the STAT6 419G mutant to IL-4, a situation similar to the observations in lymphoma cells.
In primary FL lymphoma cells, the expression of the direct STAT6 target genes FCER2, CISH, NFIL3, and CCL17 was substantially elevated without exogenous IL-4.29,30 Finally, using stably transfected STAT6 mutant and wt transfected lymphoma cells, IL-4 stimulation elicited a strong STAT6 mutant hyperactivation phenotype.
In stably transfected OCI-LY7 and OCI-LY8 lymphoma cells, mutant STAT6 proteins predominantly localized to the nucleus without a need for IL-4 stimulation or STAT6-Y641 phosphorylation. The strong effects of IL-4 stimulation on the expression of STAT6 target genes implies that ongoing nuclear-to-cytoplasmic shuttling allows access of mutant STAT6 proteins to cytoplasmic JAK kinases. Indeed, such shuttling has previously been measured.37,45 Why are mutant STAT6 proteins predominantly nuclear in localization? One possibility relates to altered interaction with chaperone proteins or nuclear import and export proteins, another to higher affinity for either DNA or protein-DNA complexes. In support of the latter, we noticed that high salt concentrations were required to extract mutated STAT6 proteins from detergent lysates of unstimulated or IL-4–stimulated transfected lymphoma cells, suggesting a firm DNA- or chromatin-bound state and, indeed, 3-D modeling data located mutated STAT6 amino acid residues in close proximity to DNA.
Why do FLs select for activating STAT6 mutations? Do these mutations elevate apoptosis thresholds or somehow facilitate survival and/or proliferation in the IL-4–rich FL microenvironment? An alternative, but not mutually exclusive hypothesis follows our observation that all STAT6-mutant FLs also carried inactivating mutations in CREBBP. This strong concordance suggests cooperation but may also imply that STAT6 mutations and CREBBP mutations compensate for each other. This is deserving of further study.
In summary, the data provided here in aggregate provide insights into the mutational activation of the IL-4/JAK/STAT6 axis in FL and provide fertile ground for future research, including but not limited to the exploration of the IL-4/JAK/STAT6 axis as a therapeutic target for small molecules (histone deacetylase inhibitors or JAK inhibitors) in FL.46-48
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the hematological malignancy group of the University of Michigan for their contribution and the genomics and bioinformatics core of the University of Michigan Comprehensive Cancer Center for providing services.
This work was supported by a gift from the Garden Fresh Company (S.N.M.), the Weatherhall foundation (M.S.K.), and a Scholar in Clinical Research Award from the Leukemia and Lymphoma Society (S.N.M.).
Authorship
Contribution: M.S.K., K.J., A.E.C., and S.N.M. enrolled patients and analyzed clinical data; M.Y., H.L., N.A.A., P.O., B.P., K.S.-C., and S.N.M. performed the laboratory research; D.B. and S.W. assisted with the structural modeling; S.J. assisted with bioinformatics analysis of next generation sequencing data; K.S. assisted with statistical methods; and S.N.M. conceived the study and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Sami N. Malek, Department of Internal Medicine, Division of Hematology and Oncology, University of Michigan, 1500 East Medical Center Drive, Ann Arbor, MI 48109-0936; e-mail: smalek@med.umich.edu.
References
Author notes
M.Y. and H.L. contributed equally to this study.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal