Key Points
HGAL protein can be myristoylated and palmitoylated, and these modifications localize HGAL to lipid rafts.
Raft localization of HGAL protein facilitates interaction with Syk, and modulation of BCR activation and signaling.
Abstract
Human germinal center–associated lymphoma (HGAL) is specifically expressed only in germinal center (GC) B lymphocytes and GC-derived lymphomas. HGAL protein decreases lymphocyte motility by inhibiting the ability of myosin to translocate actin via direct interaction with F-actin and myosin II and by activating RhoA signaling via direct interactions with RhoA-specific guanine nucleotide exchange factors. HGAL protein also regulates B-cell receptor (BCR) signaling by directly binding to and enhancing Syk kinase activity and activation of its downstream effectors. Herein we demonstrate that HGAL protein can be myristoylated and palmitoylated and that these modifications localize HGAL to cellular membrane raft microdomains with distinct consequences for BCR signaling and chemoattractant-induced cell mobility. In BCR signaling, raft localization of HGAL facilitates interaction with Syk and modulation of the BCR activation and signaling, which induces HGAL phosphorylation and redistribution from lipid raft to bulk membrane and cytoplasm, followed by degradation. In contrast, HGAL myristoylation and palmitoylation avert its inhibitory effects on chemoattractant-induced cell motility. These findings further elucidate the growing and complex role of HGAL in B-cell biology and suggest that membrane-bound and cytoplasmic HGAL protein differently regulates distinct biological processes.
Introduction
HGAL (human germinal center-associated lymphoma, also known as a germinal center-expressed transcript 2 [GCET2]) is a novel germinal center (GC)-specific gene identified by gene expression profiling.1,2 HGAL expression in subsets of diffuse large B-cell lymphoma and classic Hodgkin lymphoma patients identifies biologically distinct tumors associated with improved survival and thus may serve as a prognostic biomarker.2-5 The HGAL gene is located on chromosome 3q13 and encodes a 178–amino acid protein with 51% identity and 62% similarity to the murine GC-specific protein M17.2 Studies in M17 knockout mice revealed that this protein is dispensable for GC formation, class-switch recombination, immunoglobulin somatic hypermutation, and mounting of T-cell-dependent antibody responses.6 However, in contrast to their wild-type littermates, M17-deficient mice exhibited reduced-sized Peyer patches. Concordantly, HGAL transgenic mice generated in our laboratory exhibited increased-size intestinal Peyer patches compared with control animals and developed polyclonal B-cell lymphoid hyperplasia, hypergammaglobulinemia, and systemic reactive amyloid A (AA) amyloidosis that led to shortened survival.7 Studies in the HGAL transgenic mice as well as in-vitro studies in human lymphocytes and diffuse large B-cell lymphoma cell lines demonstrate that HGAL functions as an adaptor protein affecting several cellular functions and signaling pathways. HGAL expression decreases spontaneous and stromal cell-derived factor 1 (SDF-1)-induced or interleukin (IL)-6-induced cell motility by interacting with F-actin and myosin II and by inhibiting the ability of myosin to translocate actin.8,9 HGAL also directly interacts with the RhoA-specific guanine nucleotide exchange factors PDZ-RhoGEF and LARG. This HGAL binding stimulates the guanosine diphosphate–guanosine triphosphate exchange rate and activation of RhoA and its downstream effectors, further contributing to inhibition of cell motility.10 Furthermore, we have recently shown that HGAL enhances intracellular B-cell receptor (BCR) signaling by directly binding to Syk and by enhancing Syk kinase activity.7
To execute these various functions, HGAL protein that exhibits a hydrophilic profile needs to be localized in distinct intracellular compartments: cytoplasm for modulation of actin-myosin interaction, and cellular membrane for regulation of BCR intracellular signaling. Analysis of the HGAL protein localization by immunohistochemistry and fluorescent microscopies indeed revealed that HGAL is localized to both cell cytoplasm and membrane, despite absence of predicted transmembrane domain.7,8,11 The latter observation suggested that HGAL may be attached to cell membrane through protein interactions or posttranslational modifications. Lipid modifications such as N-myristoylation or palmitoylation are well-known methods of protein localization and stabilization control.12,13 Structural analysis of the N-terminal portion of the HGAL protein revealed the presence of putative myristoylation (1MGNS) and palmitoylation (43CFC) motifs that may mediate its membrane attachment and regulation of intracellular signaling. Herein we demonstrate that HGAL protein can be myristoylated and palmitoylated and that these protein modifications facilitate HGAL localization to membrane lipid rafts, interaction with Syk, and modulation of BCR signaling. In reverse, we also show that the localization of HGAL protein is modulated by BCR activation. Further, we demonstrate that HGAL mutations decreasing its localization to lipid rafts enhance its inhibitory effects on chemoattractant-induced cell motility.
Materials and methods
Reagents and antibodies
Mouse monoclonal anti-HGAL antibody was generated in our laboratory, as reported previously.11 Other reagents and antibodies are described in the supplemental Methods found on the Blood Web site.
Cell cultures, plasmids, transfection, and gene silencing
The non-Hodgkin lymphoma cell lines Ramos, Raji, MC116, RCK8, U2932, and VAL were cultured at 37°C and 5% carbon dioxide in RPMI 1640 medium (Fisher Scientific, Santa Clara, CA) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT), 2 mM glutamine, and 100 U/mL penicillin plus 100 μg/mL streptomycin (Invitrogen-Gibco). MCF7, HEK 293T, and HeLa cells were grown in Dulbecco’s modified Eagle medium (Invitrogen-Gibco) that was similarly supplemented with fetal bovine serum, glutamine, and penicillin/streptomycin.
The pcDNA3.1-HGAL plasmid8 was used to generate pcDNA3.1-HGAL–green fluorescent protein (GFP), HGAL-G2A, HGAL-C43A/C45A, HGAL-G2A/C43A/C45A, HGAL-G2A-GFP, HGAL-C43A/C45A-GFP, and HGAL-G2A/C43A/C45A-GFP plasmids using standard techniques. Cell transfection with plasmids and small interfering RNAs was performed using Amaxa Nucleofector kits (Amaxa, Gaithersburg, MD) as previously reported8,10 and described in the supplemental Methods.
[3H] Myristic acid and [3H] palmitic acid labeling
Raji and VAL cells (4.0 × 107) were washed twice with phosphate-buffered saline (PBS), starved for 3 hours, labeled for 6 hours in 5 mL of RPMI 1640 medium with 2 mM sodium pyruvate, 1500 μci [3H] myristic acid, and 250 μci [3H] palmitic acid, and then incubated with or without 10 ng/mL of IL-6 for 6 hours. The cells were collected, washed twice with PBS, immunoprecipitated with anti-HGAL antibody, and analyzed and visualized after separation on sodium dodecyl sulfate –polyacrylamide gel electrophoresis membranes.
Lipid raft fractionation
Cells (1.0 × 108) with or without prior treatment were fractionated following a detergent-free method adapted from Prior et al.14 Attribution of the fractions to distinct cellular localizations was performed based on detection of Lyn, transferrin receptor, and BLNK proteins that served as lipid raft, bulk membrane, and cytoplasmic protein markers, respectively. Detailed description of the methods and analyses is described in the supplemental Methods.
Western blot analysis, immunoprecipitation, chemotaxis, calcium influx, and RhoA pull-down assays
Immunofluorescence and confocal microscopy and analysis
Confocal images of the U2932, RCK8, and MCF7 cells transfected with HGAL-GFP were performed using a Nikon A1R scanning laser confocal microscope equipped with a ×60 oil immersion objective (numerical aperture of 1.4). The confocal images were acquired at acquisition rates of 4 seconds per frame. Z stacks were generated from 0.5-µm thick serial sections. All imaging measurements were carried out at 25°C. Maximum projection reconstructions from z stacks and line profile analysis were performed using Nikon NIS-Elements image analysis software. Statistical analyses of measurements derived from different cells were carried out with Microsoft Excel.
Three-dimensional analysis of colocalization of HGAL and lipid rafts was performed automatically in Volocity software (PerkinElmer) based on fluorescence intensity of each element using built-in proprietary algorithms with user feedback. Fluorescence detection threshold was set to 4 standard deviations of the noise signal for each element’s channel.16,17 To quantify changes in HGAL distribution into and out of the lipid rafts, we measured the volume (µm3) of each element and measured its “intersect” (ie, overlap) relative to its combined volume under the different conditions.18,19
Statistical analyses
For lipid raft distribution of HGAL mutants and cell motility analyses, we used the 2-tailed Student t test with unequal variances.
Results
HGAL protein is myristoylated and palmitoylated
Structural analysis of the N-terminal portion of HGAL protein revealed the presence of putative myristoylation (1MGNS) and palmitoylation (43CFC) sites which may mediate its membrane attachment and localization. Metabolic labeling with [3H] myristic acid or [3H] palmitic acid of Raji and VAL cells expressing endogenous HGAL, followed by cell lysis and immunoprecipitation with anti-HGAL antibodies, demonstrated that HGAL protein is myristoylated and palmitoylated (supplemental Figure 1), as was also shown by Pan et al in a DHL-16 lymphoma cell line transfected with exogenous HGAL.20 To demonstrate that mutagenesis of the putative HGAL myristoylation (1MGNS) and palmitoylation (43CFC) sites results in unacylated HGAL, we generated HGAL mutants G2A, C43A/C45A, and G2A/C43A/C45A, in which each of the putative lipid modification sites was mutated individually or together (Figure 1A). Metabolic labeling experiments in the HEK 293T cells expressing the wild-type or mutant HGAL proteins confirmed that HGAL is specifically myristoylated at the glycine residue in the 1MGNS motif and palmitoylated at the cysteine residues in the 43CFC motif (Figure 1B). Further, these experiments demonstrated the absence of additional motifs in the HGAL protein undergoing these lipid modifications. Because IL-6 may induce HGAL phosphorylation and localization to podosomelike structures,8 we also examined whether IL-6 stimulation for 6 hours may induce HGAL lipid modifications. Stimulation of lymphoma cells with IL-6 did not alter the extent of HGAL myristoylation and palmitoylation (supplemental Figure 1).
HGAL protein is myristoylated and palmitoylated and localizes in cellular membrane. (A) Amino acid sequences of the wild-type (WT), myristoylation and palmitoylation HGAL mutants. (B) HEK 293T cells transfected with plasmids encoding wild-type HGAL and G2A, C43A/C45A, and G2A/C43A/C45A myristoylation and palmitoylation mutants were used for metabolic labeling with [3H] myristic or [3H] palmitic acids. (C) Cellular distribution of HGAL in RCK8 (upper left image) and U2932 (upper right image) cell lines expressing WT HGAL-GFP (solid circle), G2A HGAL-GFP (solid square), C43A/C45A HGAL-GFP (open square), and G2A/C43A/C45A HGAL-GFP (open circle). HGAL distribution was determined from 3 random line profiles of a central stack of the confocal images of 12 to 15 cells. The edge of the cell was used to define the origin of the HGAL distribution and was determined by a sharp increase in the line profile. HGAL distributions were normalized to the total fluorescence from a line of 3 μm. Representative images are shown in the lower left and lower right images. Bar = 5 μm. (D) Expression of HGAL and its mutants was examined in cellular lysates using HGAL antibody and actin that was used as loading control.
HGAL protein is myristoylated and palmitoylated and localizes in cellular membrane. (A) Amino acid sequences of the wild-type (WT), myristoylation and palmitoylation HGAL mutants. (B) HEK 293T cells transfected with plasmids encoding wild-type HGAL and G2A, C43A/C45A, and G2A/C43A/C45A myristoylation and palmitoylation mutants were used for metabolic labeling with [3H] myristic or [3H] palmitic acids. (C) Cellular distribution of HGAL in RCK8 (upper left image) and U2932 (upper right image) cell lines expressing WT HGAL-GFP (solid circle), G2A HGAL-GFP (solid square), C43A/C45A HGAL-GFP (open square), and G2A/C43A/C45A HGAL-GFP (open circle). HGAL distribution was determined from 3 random line profiles of a central stack of the confocal images of 12 to 15 cells. The edge of the cell was used to define the origin of the HGAL distribution and was determined by a sharp increase in the line profile. HGAL distributions were normalized to the total fluorescence from a line of 3 μm. Representative images are shown in the lower left and lower right images. Bar = 5 μm. (D) Expression of HGAL and its mutants was examined in cellular lysates using HGAL antibody and actin that was used as loading control.
Concomitant myristoylation and palmitoylation of HGAL mediate localization to membrane lipid rafts
To examine the role of myristoylation and palmitoylation in HGAL protein localization to cellular membrane, RCK8 and U2932 cells transfected and stably expressing similar levels of GFP-tagged wild-type and G2A, C43A/C45A, and G2A/C43A/C45A HGAL mutants were examined by confocal microscopy for HGAL protein localization (Figure 1C-D). In both cell lines, wild-type HGAL was enriched in the cell membrane but was also observed to a lesser extent in the cytoplasm. Individual myristoylation (G2A) and palmitoylation (C43A/C45A) mutants demonstrated slightly decreased HGAL localization in the cell membrane in comparison with the wild-type HGAL protein. Furthermore, the G2A/C43A/C45A double mutant exhibited homogeneous HGAL distribution in cytoplasm and lost cell membrane enrichment.
Next, we used biochemical approaches to characterize localization of HGAL and its mutants in cellular membranes. Previous studies using detergent-based methods failed to demonstrate HGAL localization in lipid rafts.20 We used a detergent-free fractionation method to prepare membrane-derived vesicular fractions whose buoyant density allows flotation on a discontinuous sucrose gradient while employing a sodium carbonate buffer (pH 11) that disrupts interactions between membrane proteins and cytoplasmic complexes. Ten fractions prepared from cellular lysates of RCK8 and U2932 lymphoma cell lines stably transfected with wild-type HGAL (Figure 2A-B) were immunoblotted for HGAL, Lyn (a protein enriched in lipid rafts), transferrin receptor localized in bulk membrane, and cytoplasmic protein BLNK. Wild-type HGAL protein was enriched in membrane fractions 2-5, in which transferrin receptor and BLNK proteins were not present but which contained the lipid raft protein Lyn, suggesting specific localization of the HGAL protein in membrane lipid rafts. Wild-type HGAL protein was also detected in bulk membrane/cytoplasmic fractions (6-10) that also contained transferrin receptor and BLNK proteins. Analysis of HGAL distribution by densitometry across all the isolated cell lysate fractions revealed that a major proportion of the wild-type HGAL protein is localized in lipid rafts. Similar distribution of endogenous HGAL protein was observed in MC116, Ramos, and VAL lymphoma cell lines (supplemental Figure 2). The MCF7 breast cancer cell line was used as a model for HGAL localization driven only by its acylation, with a low probability of contributions from any interacting partners. Stable HGAL-GFP fusions in MCF7 confirmed the data derived from lymphoma cell lines (supplemental Figure 3A). Further, by using giantin as a Golgi membrane marker, we demonstrate that fractions 2-5 do not represent Golgi membrane. Overall, these findings confirm our previous microscopy findings showing HGAL protein localization in both cytoplasm and cellular membrane.8,11
HGAL myristoylation and palmitoylation modifications mediate its localization to membrane lipid rafts. Detergent-free fractionation by sucrose-density centrifugation was done on RCK8 cells (A) and U2932 cells (B) stably transfected with plasmids encoding V5-tag WT HGAL, HGAL myristoylation (G2A), HGAL palmitoylation (C43A/C45A), and HGAL palmitoylation and myristoylation (G2A/C43A/C45A) mutants. Individual fractions were immunoblotted with the indicated antibodies. Input represents 10% of the sample taken from cellular lysate after sonication and before centrifugation. Lyn, transferrin receptor, and BLNK were used as lipid raft, bulk membrane, and cytoplasm markers, respectively. HGAL was detected using V5-tag antibody. Results are representative of 3 independent experiments. (C) Distribution of HGAL in RCK8 and U2932 cells transfected with WT HGAL and HGAL mutants was generated by analyzing total densitometry in lipid raft fractions (2-5) and bulk membrane/cytoplasm fractions (6-10) using ImageJ and Origin 7 software. Data are expressed as the mean ± standard error of the mean of triplicates. P values are shown for each cell line.
HGAL myristoylation and palmitoylation modifications mediate its localization to membrane lipid rafts. Detergent-free fractionation by sucrose-density centrifugation was done on RCK8 cells (A) and U2932 cells (B) stably transfected with plasmids encoding V5-tag WT HGAL, HGAL myristoylation (G2A), HGAL palmitoylation (C43A/C45A), and HGAL palmitoylation and myristoylation (G2A/C43A/C45A) mutants. Individual fractions were immunoblotted with the indicated antibodies. Input represents 10% of the sample taken from cellular lysate after sonication and before centrifugation. Lyn, transferrin receptor, and BLNK were used as lipid raft, bulk membrane, and cytoplasm markers, respectively. HGAL was detected using V5-tag antibody. Results are representative of 3 independent experiments. (C) Distribution of HGAL in RCK8 and U2932 cells transfected with WT HGAL and HGAL mutants was generated by analyzing total densitometry in lipid raft fractions (2-5) and bulk membrane/cytoplasm fractions (6-10) using ImageJ and Origin 7 software. Data are expressed as the mean ± standard error of the mean of triplicates. P values are shown for each cell line.
We next analyzed the distribution of the G2A, C43A/C45A, and G2A/C43A/C45A HGAL mutants in membrane and cytoplasmic fractions prepared from cellular lysates of stably transfected RCK8, U2932, and MCF7 cell lines using the same fractionation method (Figure 2A-B and supplemental Figure 3B). These analyses still revealed predominant localization of the individual myristoylation and palmitoylation HGAL mutants in the lipid raft fractions, similar to the wild-type HGAL protein. Individual mutants demonstrated to varying degrees a reduction in raft preferences. For the palmitoylation-site mutant, the effect was minimal in all cases. For the myristoylation-site mutant, the degree of partial localization to the bulk membrane/cytoplasmic fractions (6-10) varied between cell lines and was modest for U2932 and MCF7. By contrast, the double mutant, eliminating both HGAL myristoylation and palmitoylation, showed a clear shift toward the bulk membrane/cytoplasmic fractions, with a statistically significant preference for localization in those fractions (P = .0016 and P = .0005 for RCK8 and U2932 cell lines, respectively). Cell treatment with methyl-β-cyclodextrin, which disrupts cholesterol-enriched lipid raft microdomains, led to redistribution of the wild-type HGAL and its mutant proteins from lipid raft fractions (2-5) to bulk membrane/cytoplasmic fractions (6-10), further confirming HGAL localization to lipid rafts (supplemental Figure 4). Overall, these biochemical studies corroborated our microscopy observations on localization of the HGAL mutants in the membrane and cytoplasm.
To confirm HGAL localization in lipid rafts, we used confocal microscopy to examine in MCF7 cells colocalization of the transiently expressed HGAL with ganglioside GM1, a widely used marker for lipid rafts (Figure 3) that is absent in the analyzed lymphoma cell lines. These studies demonstrate punctuate colocalization of HGAL with GM1 at the cellular membrane, consistent with the membrane raft localization of HGAL observed in membrane fractionation.
HGAL colocalizes in cellular membrane with lipid raft proteins. Confocal and differential interference contrast images of HGAL-GFP-expressing MCF7 cells that were cross-linked by cholera toxin B. Maximum-intensity images were generated from a series of 30 stack images acquired at a thickness of 0.5 μm. (A) HGAL channel. (B) Cholera toxin B channel. (C) Cholera toxin B overlaid on HGAL. (D) Differential interference contrast image. Results are representative of 2 independent experiments. Bar = 10 μm.
HGAL colocalizes in cellular membrane with lipid raft proteins. Confocal and differential interference contrast images of HGAL-GFP-expressing MCF7 cells that were cross-linked by cholera toxin B. Maximum-intensity images were generated from a series of 30 stack images acquired at a thickness of 0.5 μm. (A) HGAL channel. (B) Cholera toxin B channel. (C) Cholera toxin B overlaid on HGAL. (D) Differential interference contrast image. Results are representative of 2 independent experiments. Bar = 10 μm.
HGAL colocalizes with BCR complex in lipid rafts
In unstimulated B lymphocytes, the unbound BCR complex is distributed between lipid rafts and bulk membrane. However, agonist binding to BCR rapidly leads to BCR clustering in lipid rafts constitutively enriched in Lyn, resulting in activation of Syk and its downstream effectors and leading to enhanced signaling.21-24 We have previously demonstrated that HGAL enhances BCR signaling7 . In unstimulated U2932 cells, HGAL exhibits global patchy localization in cell membrane (Figure 4). After cell binding to immobilized anti-IgM F(ab′)2, HGAL relocates to the spreading cellular membrane region in contact with the anti-IgM antibodies (Figure 4 and supplemental Movies). Consequently, we next examined by biochemical fractionation experiments whether BCR and HGAL are both localized to lipid rafts (Figure 5A). Consistent with previous reports,22,24 in unstimulated Raji cells, BCR complex (represented by blotting for Igα [CD79α]) was detected in both lipid raft and bulk membrane fractions, whereas a major fraction of the HGAL protein was present in the lipid rafts, consistent with a lipid raft localization of both proteins. Furthermore, in concordance with previous reports, Igα relocated to lipid raft fractions within 5 minutes of stimulation with soluble anti-IgM F(ab′)2. By contrast, HGAL redistributed almost completely from lipid rafts to bulk membrane and cytoplasmic fractions. In whole-cell lysate, we observed a decrease in HGAL levels at 60 minutes after stimulation with soluble anti-IgM F(ab′)2 (Figure 5B). Because removal from the raft membrane compartment may make HGAL available for degradation by cytoplasmic proteasomes, we examined the effect of the proteasome inhibitor MG132 on HGAL protein levels and localization. Pretreatment with MG132 prior to anti-IgM stimulation led to stabilization of HGAL levels in cell membrane and whole-cell lysates (Figure 5A-B). For the membrane fraction, this coincides with an accumulation in lipid rafts. In total cell lysate, pSyk levels peak early after stimulation and rapidly decline, whereas total HGAL levels decrease slowly, reaching 50% of their starting level after 1 hour. This decline is blocked by MG132 and exceeds natural turnover, which is negligible in 1 hour, based on cycloheximide inhibition (Figure 5B). Overall, these observations suggested that after BCR stimulation, HGAL protein is eventually redistributed from lipid raft to bulk membrane and cytoplasm and is subsequently degraded.
HGAL relocalizes to BCR interaction membrane regions after binding to anti-IgM. U2932 cells stably expressing HGAL-GFP were pretreated with dimethylsulfoxide (control; “untreated”) or Syk inhibitor (20 nM, BAY61-3606; “Syk-inhibited”) for 30 minutes and then seeded on 8-well slides (ibidi, Verona, WI) coated with phycoerythrin-conjugated anti-human IgM F(ab′)2. Cells were fixed at 0 and 30 minutes with 3.5% paraformaldehyde and used for images as described in the “Materials and methods” section. No attachment with membrane spreading was observed on non–anti-human anti-IgM F(ab′)2-coated slides. Ig, immumoglobulin.
HGAL relocalizes to BCR interaction membrane regions after binding to anti-IgM. U2932 cells stably expressing HGAL-GFP were pretreated with dimethylsulfoxide (control; “untreated”) or Syk inhibitor (20 nM, BAY61-3606; “Syk-inhibited”) for 30 minutes and then seeded on 8-well slides (ibidi, Verona, WI) coated with phycoerythrin-conjugated anti-human IgM F(ab′)2. Cells were fixed at 0 and 30 minutes with 3.5% paraformaldehyde and used for images as described in the “Materials and methods” section. No attachment with membrane spreading was observed on non–anti-human anti-IgM F(ab′)2-coated slides. Ig, immumoglobulin.
BCR stimulation induces HGAL translocation from lipid rafts to cytoplasm and leads to HGAL degradation. (A) Raji cells were left unstimulated (“control”) or stimulated for 5 minutes in the presence or absence of MG132 (30 μM) with anti-human IgM F(ab)2 (a-IgM). Cellular lysates were subjected to detergent-free fractionation, and equal volumes of each fraction were immunoblotted with the indicated antibodies. Total densitometry in lipid raft fractions (2-5) and in bulk membrane/cytoplasm fractions (6-10) was measured, and relative distribution of HGAL and Igα was compared by randomly assigning value 1 to total densitometry measured in the lipid raft fractions (2-5). Results are representative of 3 independent experiments. (B) Left image: Raji cells were left unstimulated (“untreated”) or were stimulated with a-IgM for 15, 30, or 60 minutes. Cellular lysates were immunoblotted with HGAL, pSyk (Y525/526), or actin. Right image: Raji cells were left untreated or treated for 1hour with a-IgM with or without proteasome inhibitor MG132 (30 μM) or with cycloheximide (CHX; 40 μM) alone. Densitometry was measured for HGAL and normalized for actin content. The value 1 was assigned to the untreated sample. (C) Raji cells were left unstimulated (−) or stimulated (+) with a-IgM for 5 minutes. Membrane and cytoplasm fractionation in either PBS (pH 7.4) or sodium carbonate (Na2CO3) buffer (pH 11) was performed as described in the supplemental Materials and Methods. The fractions were immunoblotted with the indicated antibodies. HGAL (total) represents HGAL contents of whole-cell lysate before separation into cytoplasmic and membrane fractions. Membrane and cytoplasm fractions of Raji cells in sodium carbonate buffer were diluted in mild lysis buffer (MLB) and used for immunoprecipitation (IP) with monoclonal antibody to phosphotyrosine (pTyr; 4G10), followed by immunoblotting with antibody to HGAL. Densitometry before and after IgM treatment was measured and compared by assigning value 1 to the nontreated sample. Results are representative of 3 independent experiments. (D) Raji cells untreated (−) or preincubated (+) with BAY61-3606 (20 nM) for 30 minutes at 37°C were left unstimulated (−) or stimulated (+) with a-IgM for 5 minutes. Membrane and cytoplasm fractionation in sodium carbonate buffer (pH 11) was performed as in panel C, followed by immunoblotting with antibodies to HGAL, pSyk (Y525/526), and flotillin-1. Only membrane fractions are represented. SDS, sodium dodecyl sulfate.
BCR stimulation induces HGAL translocation from lipid rafts to cytoplasm and leads to HGAL degradation. (A) Raji cells were left unstimulated (“control”) or stimulated for 5 minutes in the presence or absence of MG132 (30 μM) with anti-human IgM F(ab)2 (a-IgM). Cellular lysates were subjected to detergent-free fractionation, and equal volumes of each fraction were immunoblotted with the indicated antibodies. Total densitometry in lipid raft fractions (2-5) and in bulk membrane/cytoplasm fractions (6-10) was measured, and relative distribution of HGAL and Igα was compared by randomly assigning value 1 to total densitometry measured in the lipid raft fractions (2-5). Results are representative of 3 independent experiments. (B) Left image: Raji cells were left unstimulated (“untreated”) or were stimulated with a-IgM for 15, 30, or 60 minutes. Cellular lysates were immunoblotted with HGAL, pSyk (Y525/526), or actin. Right image: Raji cells were left untreated or treated for 1hour with a-IgM with or without proteasome inhibitor MG132 (30 μM) or with cycloheximide (CHX; 40 μM) alone. Densitometry was measured for HGAL and normalized for actin content. The value 1 was assigned to the untreated sample. (C) Raji cells were left unstimulated (−) or stimulated (+) with a-IgM for 5 minutes. Membrane and cytoplasm fractionation in either PBS (pH 7.4) or sodium carbonate (Na2CO3) buffer (pH 11) was performed as described in the supplemental Materials and Methods. The fractions were immunoblotted with the indicated antibodies. HGAL (total) represents HGAL contents of whole-cell lysate before separation into cytoplasmic and membrane fractions. Membrane and cytoplasm fractions of Raji cells in sodium carbonate buffer were diluted in mild lysis buffer (MLB) and used for immunoprecipitation (IP) with monoclonal antibody to phosphotyrosine (pTyr; 4G10), followed by immunoblotting with antibody to HGAL. Densitometry before and after IgM treatment was measured and compared by assigning value 1 to the nontreated sample. Results are representative of 3 independent experiments. (D) Raji cells untreated (−) or preincubated (+) with BAY61-3606 (20 nM) for 30 minutes at 37°C were left unstimulated (−) or stimulated (+) with a-IgM for 5 minutes. Membrane and cytoplasm fractionation in sodium carbonate buffer (pH 11) was performed as in panel C, followed by immunoblotting with antibodies to HGAL, pSyk (Y525/526), and flotillin-1. Only membrane fractions are represented. SDS, sodium dodecyl sulfate.
To further elucidate this process, we used a simplified fractionation method allowing for rapid separation between membrane-attached and cytoplasmic proteins (Figure 5C and supplemental Figure 3C). Following cell lysis in PBS and in the presence of general protease inhibitors, we observed a pronounced decrease in overall HGAL levels after IgM stimulation, with the remaining HGAL representing a small and inert cytoplasmic pool. HGAL was almost entirely lost from the membrane fraction after stimulation, but the relocated HGAL pool was highly sensitive to degradation in vitro. To more effectively block degradation post lysis, we combined the rapid fractionation approach with the use of high-ionic-strength carbonate buffer to disrupt protein-protein interactions without the use of detergent. In this setting, the decrease of membrane-localized HGAL was the same, but relocalization to the cytoplasm was now evident, and overall cellular HGAL levels remained unchanged during this immediate response phase, consistent with the degradation time course in Figure 5B. These observations suggest that after BCR stimulation, HGAL is rapidly redistributed from its initial membrane raft localization to the cytoplasm. Early translocated HGAL exhibits increased sensitivity to proteases in vitro, but its cellular degradation occurs in a time-delayed manner and involves proteosomal degradation.
HGAL protein harbors a modified immunoreceptor tyrosine-based activation motif (D/EX7D/EX2YX2LX7YX2L), which is frequently used for BCR signal transduction and in which tyrosines were shown to be phosphorylated by Lyn.2,8 Consequently, we examined the presence and distribution of tyrosine-phosphorylated HGAL in cellular membrane and cytoplasm in resting cells and after BCR stimulation. To this end, we immunoprecipitated phosphotyrosine-harboring proteins in cell membrane and cytoplasm and blotted for HGAL. In unstimulated cells, phosphorylated HGAL was present mainly in the cell membrane and not in the cytoplasm (Figure 5C, right image). Shortly after BCR activation, total levels of phosphorylated HGAL increased. This increase was accompanied by a redistribution of phosphorylated HGAL from the membrane fraction to the cytoplasm.
HGAL myristoylation and palmitoylation facilitate interaction with Syk
In BCR-stimulated B lymphocytes, Syk is recruited to lipid rafts by binding to phosphorylated tyrosine residues in the immunoreceptor tyrosine-based activation motif found in the Igα/Igβ signal-transducing chains. Whether additional mechanisms may facilitate recruitment of the Syk to the vicinity of the BCR complex in lipid rafts is presently unknown. We have previously demonstrated that HGAL directly binds to Syk and increases its kinase activity.7 Inhibition of Syk with BAY61-3606 (supplemental Figure 5) did not affect HGAL movement to the membrane region in direct contact with immobilized anti-IgM antibodies (Figure 4 and supplemental Movies) but blocked the loss of HGAL from the membrane fraction (Figure 5D). We next also examined HGAL effects on Syk, because it is possible that HGAL lipid modification may play a role in HGAL binding and activation of Syk. To examine this possibility, HGAL and Syk coimmunoprecipitation experiments were performed in U2932 cells stably expressing V5-tag wild-type and G2A, C43A/C45A, and G2A/C43A/C45A HGAL mutants that were left unstimulated or stimulated with anti-IgM F(ab′)2 (Figure 6A). In the unstimulated wild-type HGAL-expressing U2932 cells, Syk coimmunoprecipitated with the HGAL protein, and the interaction increased after BCR stimulation, as was previously reported by us.7 Myristoylation-deficient G2A mutant protein demonstrated decreased coimmunoprecipitation with the Syk protein, and no interaction was observed between the palmitoylation and double-mutant HGAL proteins and Syk. We next examined the effect of these mutations on BCR activation. In accordance with our previous report,7 Syk and BLNK proteins were not phosphorylated in unstimulated cells (Figure 5B). After BCR stimulation, wild-type HGAL increased phosphorylation of Syk and BLNK, leading to enhanced calcium ion (Ca2+) influx (Figure 5B-C). In contrast, all the analyzed HGAL acylation mutants did not increase phosphorylation of Syk and BLNK. BCR stimulation–induced Ca2+ influx was also decreased in cells expressing HGAL mutants in comparison with the wild-type HGAL protein but to a different extent with each HGAL mutant. In concordance with the HGAL localization findings but in slight discrepancy with the Syk activation, single G2A and C43A/C45A mutants enhanced Ca2+ influx in comparison with the mock transfected cells but to a lesser magnitude than the wild-type HGAL protein. The double G2A/C43A/C45A HGAL mutant did not markedly affect Ca2+ influx in comparison with the mock transfected cells. Overall, these findings demonstrate that HGAL lipid modifications localizing HGAL protein to lipid rafts facilitate Syk binding and activation after BCR stimulation. However, these findings also suggest that HGAL may regulate BCR signaling, and especially Ca2+ influx, by an additional currently unknown Syk-independent mechanism.
HGAL palmitoylation and myristoylation regulates its interaction with Syk. (A) U2932 cells stably expressing WT or G2A HGAL, C43A/C45A HGAL, and G2A/C43A/C45A HGAL mutants were left unstimulated (−) or stimulated (+) with anti-human IgM F(ab′)2 (a-IgM) for 5 minutes. Whole-cell lysates were prepared, immunoprecipitated with V5-tag antibody, separated by SDS-polyacrylamide gel electrophoresis, and immunoblotted with antibodies to Syk or HGAL. Results are representative of 3 independent experiments. (B) U2932 cells stably transfected with mock vector or HGAL and its mutants were left unstimulated (“resting”) or stimulated for 1 minute with goat F(ab')2 anti-human IgM. Western blot analysis of pSyk (Y352), Syk, pBLNK (Y84), BLNK, and HGAL were performed. Actin was blotted to demonstrate equal loading. Results are representative of 3 independent experiments. (C) Kinetic analysis of calcium mobilization in U2932 cells stably transfected with HGAL or its mutants. Horizontal arrow indicates the whole process was in the presence of 1 mM EGTA; vertical arrow indicates the time points at which goat F(ab')2 anti-human IgM and calcium chloride (CaCl2) were added.
HGAL palmitoylation and myristoylation regulates its interaction with Syk. (A) U2932 cells stably expressing WT or G2A HGAL, C43A/C45A HGAL, and G2A/C43A/C45A HGAL mutants were left unstimulated (−) or stimulated (+) with anti-human IgM F(ab′)2 (a-IgM) for 5 minutes. Whole-cell lysates were prepared, immunoprecipitated with V5-tag antibody, separated by SDS-polyacrylamide gel electrophoresis, and immunoblotted with antibodies to Syk or HGAL. Results are representative of 3 independent experiments. (B) U2932 cells stably transfected with mock vector or HGAL and its mutants were left unstimulated (“resting”) or stimulated for 1 minute with goat F(ab')2 anti-human IgM. Western blot analysis of pSyk (Y352), Syk, pBLNK (Y84), BLNK, and HGAL were performed. Actin was blotted to demonstrate equal loading. Results are representative of 3 independent experiments. (C) Kinetic analysis of calcium mobilization in U2932 cells stably transfected with HGAL or its mutants. Horizontal arrow indicates the whole process was in the presence of 1 mM EGTA; vertical arrow indicates the time points at which goat F(ab')2 anti-human IgM and calcium chloride (CaCl2) were added.
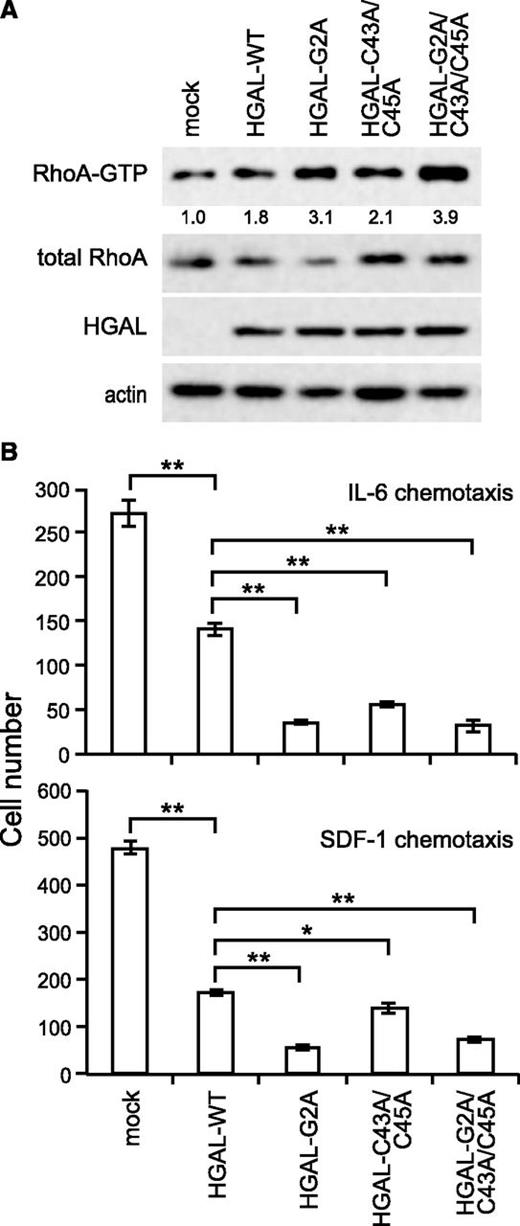
RhoA activation and cell motility inhibition are augmented by the nonmyristoylated and nonpalmitoylated HGAL
Our previous studies have shown that HGAL decreases spontaneous and chemoattractant-induced cell motility by activation of the RhoA signaling pathway and by direct interaction with actin and myosin.8-10 RhoA activation after exposure to fibronectin was enhanced in RCK8 cells expressing G2A, C43A/C45A, and G2A/C43A/C45A HGAL mutants compared with wild-type controls but to a different extent (Figure 7A). Concordantly, RCK8 cells expressing G2A, C43A/C45A, and G2A/C43A/C45A HGAL mutants showed a further significant reduction in cell motility in response to IL-6 (P < .00001, P < .0001, and P < .00001, respectively) and to SDF-1 (P < .00001, P < .05, and P < .00001, respectively) in comparison with the inhibitory effect observed in the wild-type HGAL vs mock expressing cells (Figure 7B).
HGAL palmitoylation and myristoylation mutants enhance RhoA activation and greatly inhibit cell motility. (A) RCK8 cells stably expressing WT and G2A, C43A/C45A, and G2A/C43A/C45A HGAL mutants were starved for 8 hours and then seeded on fibronectin for 60 minutes. Cellular extracts were prepared and RhoA pull-down assay was performed. Equal loading was confirmed by immunoblotting with actin antibodies. Results are representative of 3 independent experiments. Densitometry analysis of normalized RhoA-GTP to total RhoA is presented. (B) RCK8 cells stably expressing mock vector or WT, G2A, C43A/C45A, and G2A/C43A/C45A HGAL mutants were used for IL-6 or SDF-1 chemotaxis assay performed in triplicate. Data are expressed as the mean ± standard error of the mean of triplicates. *P < .05; **P < .001. Results are representative of 2 independent experiments. GTP, guanosine triphosphate.
HGAL palmitoylation and myristoylation mutants enhance RhoA activation and greatly inhibit cell motility. (A) RCK8 cells stably expressing WT and G2A, C43A/C45A, and G2A/C43A/C45A HGAL mutants were starved for 8 hours and then seeded on fibronectin for 60 minutes. Cellular extracts were prepared and RhoA pull-down assay was performed. Equal loading was confirmed by immunoblotting with actin antibodies. Results are representative of 3 independent experiments. Densitometry analysis of normalized RhoA-GTP to total RhoA is presented. (B) RCK8 cells stably expressing mock vector or WT, G2A, C43A/C45A, and G2A/C43A/C45A HGAL mutants were used for IL-6 or SDF-1 chemotaxis assay performed in triplicate. Data are expressed as the mean ± standard error of the mean of triplicates. *P < .05; **P < .001. Results are representative of 2 independent experiments. GTP, guanosine triphosphate.
Discussion
Protein-lipid modifications are implicated in the process of protein trafficking between organelles, in the segregation or clustering of proteins in membrane compartments, and in protein stability and function. N-myristoylation is the covalent addition of the fatty acid myristate to an N-terminal glycine residue via an amide linkage after the removal of the N-terminal methionine. Palmitoylation refers to the addition of palmitate to a cysteine residue, either at the C or N terminus. Herein, we demonstrate that HGAL, a GC-specific protein, undergoes myristoylation and palmitoylation that promote its localization to membrane lipid rafts. This localization facilitates Syk activation and regulation of BCR signaling while ameliorating its effects on RhoA activation and cell motility. Although our findings confirm the observation by Pan et al20 on HGAL lipid modifications, they extend Pan et al’s findings by demonstrating the specificity of the lipid modification motifs, the localization of modified HGAL to lipid rafts, and their functional significance. Although the consensus motifs for myristoylation and palmitoylation are well recognized, there are many proteins for which the palmitoylated cysteine residues are not associated with a defined consensus sequence. Herein we show that the identified consensus motifs are essential for the palmitoylation and myristoylation of HGAL and that they are the only HGAL motifs undergoing these lipid modifications.
Our data also show that these lipid modifications are necessary to promote HGAL protein localization in membrane lipid rafts. The raft localization of soluble proteins with conditional membrane localization capability is very sensitive to disruption, and earlier studies by Pan et al20 had used a Triton-X100-based purification scheme. In this detergent-based analysis, HGAL did not localize to raft fractions but shared this behavior with Lyn, a well-established marker of raft fractions. By contrast, our detergent-free fractionation approach cofractionated both Lyn and HGAL and separated them from nonraft markers. Furthermore, this fractionation behavior was sensitive to cholesterol sequestration by methyl-β-cyclodextrin, a well-established raft disruption reagent, and the fraction scheme reproduced the activation-dependent relocalization of BCR. We further confirmed the biochemically determined HGAL localization in lipid rafts by confocal microscopy studies in which HGAL colocalized with the known lipid raft component GM1. We also show that mutations eliminating consensus motifs for HGAL lipid modifications lead to HGAL redistribution away from the lipid rafts. Moreover, these mutations affect HGAL function, suggesting that HGAL localization in cellular membrane lipid rafts is important for mediating its effects on BCR signaling. However, we demonstrate that each HGAL lipid modification partially (and to a different extent) averts its inhibitory effects on cell motility. These observations suggest that lipid-modified and nonmodified cellular fractions of the HGAL protein play distinct roles in regulation of different cellular biological processes.
Similar to Lyn, which was shown to phosphorylate HGAL, a fraction of the HGAL protein pool is colocalized with BCR to the lipid rafts of unstimulated B-cell lymphoma cells. Syk plays a critical role in signal transmission but does not undergo lipid modification. HGAL localization to rafts and interaction with Syk may therefore serve to enhance the recruitment of Syk to the BCR signaling complex upon activation. However, the processing of BCR and HGAL after stimulation is very distinct. After ligand-induced cross-linking, BCR is initially recruited to lipid rafts before subsequent internalization into early endosomes.23 By contrast, the activation-sensitive portion of HGAL already resides in rafts and is rapidly removed from the lipid rafts to bulk membrane and cytoplasm. In the cytoplasm, activated HGAL undergoes degradation in a time-delayed fashion. This rapid change in HGAL localization suggests that HGAL might exert most of its enhancement effects on Syk activation in the early stages of stimulation (5-10 minutes). This finding is in concordance with our published observations.7 Furthermore, much of the HGAL relocalization is significantly faster than the endocytosis of BCR, which is known to preferentially follow clathrin-dependent endocytosis. A decrease in the strong HGAL signal in both the raft and bulk fractions suggests that HGAL may have alternative routes available for its removal from the plasma membrane, including direct transfer to the cytoplasm. In addition, the qualitative impact of mutations in the different acylation sites is consistent across assays, but quantitative differences exist for the relative impact of specific mutations on 3 types of readouts: activation of Syk and BLNK, binding of Syk, and Ca2+ influx and mobility. These differences raise the possibility that HGAL may regulate BCR signaling by other, currently unknown Syk-independent mechanisms. Further studies to examine these hypotheses are needed.
Our findings also show that HGAL localization in cellular membrane lipid rafts is required for proper binding to Syk and for subsequent Syk activation. HGAL lipid mutants, especially palmitoylation mutants and dual myristoylation and palmitoylation mutants, exhibit no association with Syk and consequently do not enhance activation of Syk and BLNK. Overall, these findings demonstrate the importance of temporal-spatial HGAL localization in lipid rafts for regulation of BCR signaling. We note that compared with the fully acylated protein, the nonacylated HGAL mutant does not show enrichment in or near the plasma membrane. This change in cellular distribution is associated with loss of membrane-specific HGAL functions, such as enhancement of BCR signaling. These findings also suggest that either modification may be sufficient to achieve general raft localization and residual ability to bind to Syk for palmitoylated HGAL, but the ability to shuttle between different membrane compartments in a manner that facilitates HGAL functions may require both modifications. Further studies are necessary to examine the precise mechanism of HGAL-induced Syk activation. Because HGAL and Lyn colocalize in lipid rafts, it is possible that HGAL may modulate Lyn binding to Syk or enhance Syk activation by Lyn. Studies addressing these questions are in progress in our laboratory.
HGAL was previously shown to bind directly to actin and myosin and to modulate interactions between these proteins.9 Further, we have previously shown that HGAL may increase actin polymerization by activation of the RhoA signaling pathway,10 whereas Syk may bind to actin and regulate cytoskeleton.25 Actin may play an important role in regulation of membrane mobility, BCR signaling, and antigen internalization.26-28 Previous studies demonstrated that BCR stimulation leads to a rapid actin depolymerization, breaking down membrane diffusion barriers and allowing ligand-clustered BCR complexes and lipid raft coalescence.28 Lipid raft–localized HGAL may contribute to actin polymerization and membrane barrier formation, whereas rapid HGAL exit from lipid rafts after BCR activation may facilitate actin depolymerization and the formation of polarized large lipid raft clusters.
Antigen encounter by BCR is important for GC reaction, during which HGAL is expressed in GC lymphocytes and initiates two critical processes: signal transduction and antigen selection and presentation. These processes are needed for efficient selection of antigen-specific GC cells that will differentiate into memory and plasma cells. HGAL may contribute to the GC process by augmenting BCR signaling and restricting cell exit from the GCs by decreasing cell motility. Whether HGAL also plays a role in antigen internalization, processing, and presentation is currently under investigation in our laboratory. Studies addressing the potential role of HGAL in controlling lipid raft–linked BCR signaling, antigen internalization, and cytoskeleton regulation will elucidate the growing role of HGAL in B-cell biology.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work is supported by National Institutes of Health, National Cancer Institute grants CA109335 (I.S.L.) and CA9888-1 (R.L.) and National Institute of Diabetes and Digestive and Kidney Diseases grant DK097194 (M.H.A.), and by Bankhead-Coley Cancer Research Program grant 1BD-09 (R.S.). I.S.L is supported by the Lymphoma Research Foundation and the Dwoskin Family, Recio Family, and Anthony Rizzo Family Foundations. M.H.A. is supported by the Diabetes Research Institute Foundation.
Authorship
Contribution: X.J., V.T.M., R.L., and I.S.L. designed the studies; X.L., R.S., X.J., J.N.S., G.M., and V.T.M. performed the experiments and analyzed the data; M.A., R.L., and I.S.L. analyzed the data; and V.T.M., R.L., and I.S.L. wrote the paper. All authors reviewed the manuscript and agree with its content.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Izidore S. Lossos, Division of Hematology-Oncology, Sylvester Comprehensive Cancer Center, University of Miami, 1475 NW 12th Ave (D8-4), Miami, FL 33136; e-mail: ilossos@med.miami.edu.
References
Author notes
X.L., R.S., and X.J. contributed equally to this study as first authors.
R.L. and I.S.L. contributed equally as senior authors.

![Figure 1. HGAL protein is myristoylated and palmitoylated and localizes in cellular membrane. (A) Amino acid sequences of the wild-type (WT), myristoylation and palmitoylation HGAL mutants. (B) HEK 293T cells transfected with plasmids encoding wild-type HGAL and G2A, C43A/C45A, and G2A/C43A/C45A myristoylation and palmitoylation mutants were used for metabolic labeling with [3H] myristic or [3H] palmitic acids. (C) Cellular distribution of HGAL in RCK8 (upper left image) and U2932 (upper right image) cell lines expressing WT HGAL-GFP (solid circle), G2A HGAL-GFP (solid square), C43A/C45A HGAL-GFP (open square), and G2A/C43A/C45A HGAL-GFP (open circle). HGAL distribution was determined from 3 random line profiles of a central stack of the confocal images of 12 to 15 cells. The edge of the cell was used to define the origin of the HGAL distribution and was determined by a sharp increase in the line profile. HGAL distributions were normalized to the total fluorescence from a line of 3 μm. Representative images are shown in the lower left and lower right images. Bar = 5 μm. (D) Expression of HGAL and its mutants was examined in cellular lysates using HGAL antibody and actin that was used as loading control.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/125/4/10.1182_blood-2014-04-571331/4/m_649f1.jpeg?Expires=1765893596&Signature=EQZbBLlM9EoJIn7zKF97XNF-G5iN-R8qwycLuIQDa7vXux~Xxe30o~dRezKkjT8rUcOCDBk26zzMXmLfE8YOhSRUpfxn63gHRFfgongSsdpqPXUkvgyhjAIK9xV744dgmx5-RJLSizf8AdM-nje-jlCyNnkMU71b2v8Ha4mlPfOIdifStBRCt7Jlb2xj49SFXmVIuosKDEwgd7BW8v0DSaHifyCweRfGvZFb5g3uo8qfVUAyI0aSMRUr2Epz2R8CYQwO4H9ZGjYC2VxETwcqzPwi1x73RxFIvJY0kTKIvySP9Z2ndYw1O~8swI~mQ8-CXclSjXXn8zsay3eFFGfQ5w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)