Key Points
miR-18b overexpression identified patients with poor prognosis in 2 large prospective homogenously treated MCL cohorts.
miR-18b overexpression adds prognostic information to the MIPI-B prognosticator.
Abstract
Recent studies show that mantle cell lymphoma (MCL) express aberrant microRNA (miRNA) profiles; however, the clinical effect of miRNA expression has not previously been examined and validated in large prospective homogenously treated cohorts. We performed genome-wide miRNA microarray profiling of 74 diagnostic MCL samples from the Nordic MCL2 trial (screening cohort). Prognostic miRNAs were validated in diagnostic MCL samples from 94 patients of the independent Nordic MCL3 trial (validation cohort). Three miRNAs (miR-18b, miR-92a, and miR-378d) were significantly differentially expressed in patients who died of MCL in both cohorts. MiR-18b was superior to miR-92a and miR-378d in predicting high risk. Thus, we generated a new biological MCL International Prognostic Index (MIPI-B)-miR prognosticator, combining expression levels of miR-18b with MIPI-B data. Compared to the MIPI-B, this prognosticator improved identification of high-risk patients with regard to cause-specific, overall, and progression-free survival. Transfection of 2 MCL cell lines with miR-18b decreased their proliferation rate without inducing apoptosis, suggesting that miR-18b may render MCL cells resistant to chemotherapy by decelerating cell proliferation. We conclude that overexpression of miR-18b identifies patients with poor prognosis in 2 large prospective MCL cohorts and adds prognostic information to the MIPI-B. MiR-18b may reduce the proliferation rate of MCL cells as a mechanism of chemoresistance.
Introduction
Mantle cell lymphoma (MCL) is an aggressive B-cell lymphoma subtype characterized by the pathogenic hallmark, the t(11;14)(q13;q32) translocation, which leads to overexpression of the cell cycle–promoting protein, cyclin D1.1 New treatment regimens, including CD20 antibodies, high-dose chemotherapy, and autologous stem cell transplantation (ASCT), have improved survival exceptionally.2-4 Despite this, long-term follow-up data from the second Nordic Lymphoma Group MCL Trial cohort (MCL2) show that 57% of younger MCL patients (≤65 years) relapse within a 10-year period.5 Risk stratification is therefore of great importance in identifying patients who are eligible for novel or alternative treatment regimens. The current prognostic models, the clinical MCL International Prognostic Index (MIPI)6 and the biological MIPI (MIPI-B) that includes staining with the proliferation marker Ki-67, assign patients to a high, intermediate, or low risk group. This prognosticator has been investigated in several cohorts4,7-11 and was useful in most.7-10 The MIPI-B, however, does not identify specific biological pathways designating aggressive subtypes of MCL.
MicroRNAs (miRNAs) are a class of noncoding RNAs that inhibit translation of specific messenger RNAs and can function as both tumor suppressor miRNAs and oncogenic miRNAs depending on the cellular context.12 Because of their small size (19-22 nucleotides), miRNAs are well preserved and can be successfully examined in formalin-fixed paraffin-embedded (FFPE) tissue, showing expression levels that correlate with those obtained in cryopreserved tissue.13 We have previously purified miRNAs from FFPE tissue in cutaneous T-cell lymphoma14 and diffuse large B-cell lymphoma15 and obtained reproducible miRNA levels in samples stored for up to 30 years.
Over the last 5 years, several studies have shown that miRNAs delineate aberrant molecular pathways and predict survival in MCL patients.16,17 Furthermore, the knockdown of specific miRNAs in 2 MCL mouse models has demonstrated efficacy in inhibiting lymphoma growth.18,19 However, there has been only modest consistency between clinical studies, and, at this point, miRNA expression in MCL has not yet been examined in large prospective uniformly treated patient cohorts.
To identify aberrant biological pathways and find prognostic biomarkers, we examined miRNA expression levels in 168 MCL patients from almost-identical trial cohorts, the MCL2 and the third Nordic Lymphoma Group MCL Trial (MCL3), according to the Reporting Recommendations for Tumor Marker Prognostic Studies guidelines.20 All patients had been treated with high-dose immunochemotherapy and ASCT, and had long-term follow-up. We initially analyzed the genome-wide miRNA expression profile of diagnostic patient samples in the MCL2 cohort and linked these findings with comprehensive clinical follow-up data. Validation of differentially expressed miRNAs was performed in samples from patients in the MCL3 cohort to determine the prognostic value of miRNA expression in MCL patients. Finally, the functional implications of aberrant miRNA expression were studied in MCL cell lines.
Methods
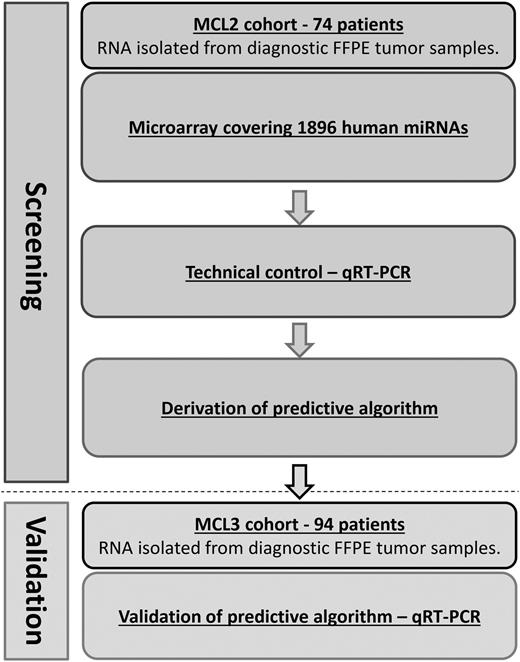
Figure 1 provides a flowchart summarizing the experimental procedures.
Flowchart of study. A dual-method approach consisting of an initial screening by genome-wide miRNA microarray of MCL samples from the MCL2 cohort, with subsequent confirmation of differentially expressed miRNAs by qRT-PCR in the same patients. Confirmed differentially expressed miRNAs identified in the MCL2 cohort were validated by qRT-PCR in samples from the MCL3 cohort. qRT-PCR, quantitative reverse-transcription polymerase chain reaction.
Flowchart of study. A dual-method approach consisting of an initial screening by genome-wide miRNA microarray of MCL samples from the MCL2 cohort, with subsequent confirmation of differentially expressed miRNAs by qRT-PCR in the same patients. Confirmed differentially expressed miRNAs identified in the MCL2 cohort were validated by qRT-PCR in samples from the MCL3 cohort. qRT-PCR, quantitative reverse-transcription polymerase chain reaction.
Study population
All patients included in this study were enrolled in 1 of 2 similar prospective trial cohorts (MCL2 or MCL3), the study designs of which have been published previously.2,21 Briefly, both cohorts consisted of newly diagnosed, previously untreated stage II-IV MCL patients <66 years old. All diagnostic specimens were centrally reviewed by at least 1 member of the central Nordic MCL pathology board, and the diagnosis was confirmed according to World Health Organization criteria.22 All patients were followed with a complete workup including clinical history, physical examination, blood and bone marrow examination, and computed tomography or positron emission tomography–computed tomography scans on study entry and, subsequently, every 6 months for 5 years. After 5 years, patients were followed with annual clinical assessment, standard blood tests, and minimal residual disease (MRD) analysis.23
Both cohorts received the same treatment regimens consisting of rituximab (R)–maxi–CHOP (cyclophosphamide, hydroxy doxorubicin, vincristine, and prednisone) alternating with R-high-dose cytarabine (6 cycles total), followed by high-dose BEAM/C (bis-chloroethylnitrosourea, etoposide, cytarabine, and melphalan or cyclophosphamide) and ASCT. The only difference between the 2 regimens was that MCL3 patients with unconfirmed complete remission or partial remission received 90Y-ibritumomab tiuxetan in addition to BEAM or carmustine/etoposide/cytarabine before ASCT. The addition of 90Y-ibritumomab tiuxetan did not have any impact on treatment outcome or adverse events, and overall survival (OS), event-free survival, and time to progression were similar in the 2 cohorts.21 In both cohorts, a fraction of these patients also received preemptive rituximab at molecular relapse during follow-up.23
Patient samples
A total of 172 FFPE preserved diagnostic MCL biopsy samples from 172 MCL patients from both cohorts (76 from MCL2 and 96 from MCL3) were retrieved from the respective Nordic MCL pathology board. Four patients were excluded due to poor quality of the RNA extracted from the MCL specimens. All included patient samples had confirmed overexpression of cyclin D1 or carried the t(11;14)(q13;q32) translocation. Ki-67 expression was evaluated by immunohistochemistry in 145 patient samples, and a marker for MRD23 was available in 96 of these.
The clinical trials (MCL2: ISRCTN 8786 6680 and MCL3: NCT 00514475) were conducted in accordance with the Declaration of Helsinki and approved by the relevant ethics committees of all Nordic countries. Informed consent was obtained from all participants.
RNA extraction from FFPE tissue
Total RNA from all samples was isolated from 4 10-μm tissue sections using the RecoverAll Total Nucleic Acid Isolation Kit (Ambion, Carlsbad, CA) according to the manufacturer’s guidelines. The samples were incubated in xylene at 50°C to remove paraffin, followed by ethanol wash. Proteins were degraded by digestion buffer and protease at 50°C and 80°C, respectively. Subsequently, isolation buffer and ethanol were added, samples were bound to a spin column, and DNA was degraded by DNase treatment. The filter was washed, and total RNA was eluted in a 60-μL elution solution. Total RNA quantity (optical density [OD] 260 nm) and quality (260/280 ratio) were measured by an ND-1000 Spectrophotometer (Thermo Scientific, Wilmington, DE).
miRNA microarray analysis
For miRNA microarray profiling, 500 ng of total RNA from samples or reference (AM6000 [common RNA pool for identifying a maximum number of expressed miRNAs in patient samples]; Ambion) was labeled with Hy3 and Hy5 fluorescent dyes, respectively, using the miRCURY LNA Array Power labeling kit (Exiqon A/S, Vedbæk, Denmark). Samples with RNA concentrations lower than 160 ng/μL were vacuum centrifuged (SpeedVac) to obtain higher concentrations. All samples were labeled the same day with the same master mix to minimize technical variation.
The labeled samples were hybridized to miRCURY LNA seventh-generation arrays (v.2.2; Exiqon A/S) containing capture probes targeting all human miRNAs registered in miRBASE release 18.0 (1896 human miRNAs). The hybridization was performed overnight at 56°C according to the manufacturer’s specifications using an HS 4800 hybridization station (Tecan, Männedorf, Switzerland). Because it was not possible to hybridize all arrays at the same time point, samples were randomly split into 6 batches to minimize day-to-day variation in the hybridization process and analyzed over 6 consecutive days. After hybridization, the microarray slides were scanned in an ozone-free environment to prevent potential bleaching of the fluorescent Hy5 dye. Scanning was done using a G2565BA Microarray Scanner System (Agilent Technologies, Santa Clara, CA) at 10-μm resolution, and the resulting images in tag image file format (TIFF) were analyzed using the GenePix 6.1 software on standard settings.
Microarray data processing
Processed microarray image data (GenePix median spot intensities) were read into the R statistical environment and preprocessed using the limma package.24 The arrays were background corrected using the “normexp” method with an offset of 10 to account for spatial biases as suggested by Ritchie et al,25 and between-array variance was eliminated by quantile normalization.26,27 Expression profiles and clinical variables were subjected to principal component analysis (singular-value decomposition) to assess whether data were affected by unexpected confounding effects.
A differential expression analysis of the foreground intensities was conducted for the cause-specific death contrast. Empirical Bayes moderated statistics were assessed and adjusted for multiple testing using the Benjamini-Hochberg method.
Quality assessment of patient samples
Principal components analysis of miRNA microarray data and sample-related parameters (anatomical site of origin of MCL biopsy, country, batch, SpeedVac procedure, cytology, or growth pattern) confirmed that none of these factors had any influence on the quality of the microarray data (data not shown).
miRNA qRT-PCR
Ten nanograms of RNA were reverse transcribed in 10-μL reactions using miRCURY LNA Universal RT miRNA PCR and Polyadenylation and cDNA Synthesis Kit (Exiqon A/S). Complementary DNA (cDNA) was diluted 40 times and assayed in duplicate 10-μL PCR assays according to protocol. Negative controls excluding template from the RT reaction were performed and profiled in parallel. The qRT-PCR reactions were performed on a LightCycler 480 II instrument (Roche, Basel, Switzerland) in 96- and 384-well plates. The amplification curves were analyzed using Roche LightCycler v.1.5.1.62 software for determination of crossing point (Cp; by the second derivative method) and for melting curve analysis. All assays were inspected for distinct and uniform melting curves to verify that the correct amplicon was amplified in all samples. Furthermore, samples were included in the data analysis only when their Cp value was 3 less than the negative control, and with Cp <39. Samples that did not pass these criteria were omitted from further analysis. To eliminate bias of miRNA concentrations due to differences in initial RNA concentration, data were normalized using averaging windows (loess method). MiR-500a-5p, miR-335-3p, and miR-423-5p had the lowest variance among expressed miRNAs on the microarray, which was confirmed by qRT-PCR, and were selected as reference miRNAs.
Statistics and response criteria
Differences in baseline characteristics between the 2 cohorts were calculated using the Pearson χ2 test and the Mann-Whitney nonparametric test. Outcome-related variables were assessed with Kaplan-Meier analyses and compared using the log-rank test. The prognostic information of the miRNAs was evaluated using the minimal-redundancy-maximal-relevance criterion.26,27 Comparison of the MIPI-B prognosticator and the MIPI-B-miR prognosticator was performed with respect to the prediction of high-risk groups. Statistical analyses were performed using SPSS v.20.0.0 (IBM, New York, NY) and R v.2.1.4 (www.r-project.org). Two-sided tests with a significance level of 5% were used for all analyses.
The prospective nature of the 2 MCL trials enabled us to use cause-specific survival (CSS) as the primary end point according to response criteria of the International Workshop.28 Progression-free survival (PFS) and OS were used as secondary end points. Multivariate Cox regression analysis was performed with backward selection to assess prognostic factors.
Cell culture
The MCL cell line Z-138 was obtained from American Type Culture Collection, whereas Jeko-1 was provided by the European MCL Network. The cells were maintained as suspension cultures in either RPMI 1640 (Jeko-1) or Iscove modified Dulbecco medium (Z-138) supplemented with 20% heat-inactivated fetal calf serum, 10 U/mL penicillin, and 10 μg/mL streptomycin. All tissue culture reagents were purchased from Life Technologies. The cells were maintained in a humidified chamber with 5% carbon dioxide at 37°C.
Electroporation
Jeko-1 and Z-138 cells were transfected in a Nucleofector II Device (Lonza) using program X-001. Briefly, 5 × 106 cells were resuspended in 100 μL of Solution V (Lonza) containing 1 μM of either miR-18b mirVana miRNA mimic (Life Technologies) or a scrambled negative control mirVana miRNA mimic (Life Technologies). Immediately after electroporation, the cells were transferred to preheated complete medium and incubated at 37°C until further analysis by flow cytometry (FACSCalibur, Becton Dickinson) or XTT (2,3-bis[2-methoxy- 4-nitro-sulfophenyl]-2H-tetrazolium-5-carboxanilide) assay.
Apoptosis and cell proliferation/viability assays
Apoptosis was assessed using the Annexin V-PE/7-AAD Apoptosis Detection Kit (BD Biosciences) according to the manufacturer’s protocol. Briefly, 24 hours after transfection, 1 × 105 cells were incubated with 5 μL of Annexin V-PE and 5 μL of 7-AAD in 100 μL of 1 × Annexin V Binding Buffer at room temperature. After incubating for 15 minutes, 400 μL of 1 × Annexin V Binding Buffer was added, and the apoptotic cells were then analyzed by flow cytometry.
To study cell proliferation, Jeko-1 and Z-138 cells were seeded in 96-well plates immediately after transfection at a density of 5 to 10 × 104 cells per well in 50 and 100 μL of complete medium, respectively. Cell number was determined 0 to 8 days after transfection by adding 25 or 50 μL of XTT (Roche Diagnostics) reagent to each well and measuring the absorbance at 450 nm (reference 650 nm) after 3 hours of incubation at 37°C.
Results
Clinical characteristics of the study cohorts
Baseline characteristics of MCL2 and MCL3 patients were similar with respect to gender, initial duration of response, World Health Organization performance status, white blood cell count, lactate dehydrogenase, age, Ki67 expression, and MIPI score but differed significantly with regard to cytological variant. Follow-up was complete for all patients of both cohorts, with a median follow-up of 6.4 years for MCL2 and 3.7 years for MCL3 (see supplemental Table 1, available on the Blood Web site).
miRNA microarray screening of the MCL2 cohort reveals 17 differentially expressed miRNAs in patients who died of MCL
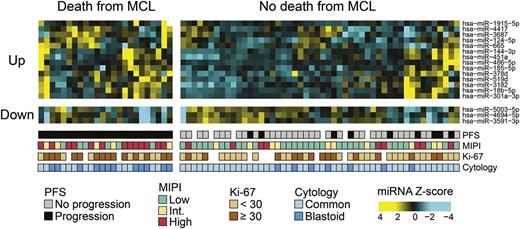
We initially assessed the global miRNA profile of 74 diagnostic samples from patients of the MCL2 cohort by miRNA microarray. To identify prognostic miRNAs in MCL, each microarray was subjected to a statistical hypothesis test (see “Methods”) with regard to the CSS. This end point was chosen to evaluate the impact of the specific biological (miRNA) aberrations and was possible because of the extensive follow-up in the 2 clinical trials. Seventeen miRNAs were found to be differentially expressed (adjusted P values <.05; see Table 1), with 14 miRNAs being upregulated and 3 miRNAs being downregulated in patients who died of MCL. A heatmap of the differentially expressed miRNAs is shown in Figure 2.
Significantly differentially expressed miRNAs according to cause-specific survival
| miRNA . | MCL2 (screening cohort) . | MCL3 (validation cohort) . | |
|---|---|---|---|
| Microarray . | qRT-PCR . | qRT-PCR . | |
| hsa-miR-124-5p | .0002 | — | — |
| hsa-miR-4417 | .0002 | .02 | .2309 |
| hsa-miR-18b-5p | .0004 | .02 | .0002 |
| hsa-miR-3687 | .0046 | .01 | .1722 |
| hsa-miR-519d | .0046 | — | — |
| hsa-miR-378d | .0092 | .02 | .0137 |
| hsa-miR-144-3p | .0173 | .03 | .142 |
| hsa-miR-486-5p | .0173 | .05 | .0644 |
| hsa-miR-1915-5p | .0176 | — | — |
| hsa-miR-665 | .0259 | — | — |
| hsa-miR-185-5p | .0308 | .04 | .059 |
| hsa-miR-4694-5p | .0405 | — | — |
| hsa-miR-5003-5p | .0427 | — | — |
| hsa-miR-301a-3p | .0439 | .43 | — |
| hsa-miR-3182 | .0439 | — | — |
| hsa-miR-3591-3p | .0463 | — | — |
| hsa-miR-451a | .0496 | .05 | .4075 |
| hsa-miR-92a-3p | .0842 | .04 | .005 |
| hsa-miR-146a-5p | .0997 | .16 | .6801 |
| miRNA . | MCL2 (screening cohort) . | MCL3 (validation cohort) . | |
|---|---|---|---|
| Microarray . | qRT-PCR . | qRT-PCR . | |
| hsa-miR-124-5p | .0002 | — | — |
| hsa-miR-4417 | .0002 | .02 | .2309 |
| hsa-miR-18b-5p | .0004 | .02 | .0002 |
| hsa-miR-3687 | .0046 | .01 | .1722 |
| hsa-miR-519d | .0046 | — | — |
| hsa-miR-378d | .0092 | .02 | .0137 |
| hsa-miR-144-3p | .0173 | .03 | .142 |
| hsa-miR-486-5p | .0173 | .05 | .0644 |
| hsa-miR-1915-5p | .0176 | — | — |
| hsa-miR-665 | .0259 | — | — |
| hsa-miR-185-5p | .0308 | .04 | .059 |
| hsa-miR-4694-5p | .0405 | — | — |
| hsa-miR-5003-5p | .0427 | — | — |
| hsa-miR-301a-3p | .0439 | .43 | — |
| hsa-miR-3182 | .0439 | — | — |
| hsa-miR-3591-3p | .0463 | — | — |
| hsa-miR-451a | .0496 | .05 | .4075 |
| hsa-miR-92a-3p | .0842 | .04 | .005 |
| hsa-miR-146a-5p | .0997 | .16 | .6801 |
Values represent adjusted P values. Dashes indicate that analysis was not performed. The miRNAs in bold are significantly differently expressed in both the screening cohort and the validation cohort; the miRNAs in italics are downregulated in patients who die of MCL; and the miRNAs in normal typeface are upregulated in patients who die of MCL.
Hsa, Homo sapiens.
Heatmap of miRNAs differentially expressed in patients who die of MCL. Hierarchical clustering of MCL samples from patients of the MCL2 cohort. The microarray-generated heatmap demonstrates differentially expressed miRNAs (adjusted P value <.05) in patients who died of MCL (cause-specific death) compared with patients who did not die of MCL. Seventeen miRNAs were found to be differentially expressed; of these, the 14 miRNAs shown at the top part of the figure (Up) had higher expression in the patients who died of MCL, whereas the 3 miRNAs shown in the bottom part (Down) had lower expression in the patients who died of MCL. Int., intermediate.
Heatmap of miRNAs differentially expressed in patients who die of MCL. Hierarchical clustering of MCL samples from patients of the MCL2 cohort. The microarray-generated heatmap demonstrates differentially expressed miRNAs (adjusted P value <.05) in patients who died of MCL (cause-specific death) compared with patients who did not die of MCL. Seventeen miRNAs were found to be differentially expressed; of these, the 14 miRNAs shown at the top part of the figure (Up) had higher expression in the patients who died of MCL, whereas the 3 miRNAs shown in the bottom part (Down) had lower expression in the patients who died of MCL. Int., intermediate.
qRT-PCR confirms miRNA microarray findings in the MCL2 cohort
To confirm the microarray findings, we chose 26 miRNAs to be analyzed by qRT-PCR in the same 74 MCL2 patient samples: 17 miRNAs were selected on the basis of adjusted P values <.05 on the microarray (Table 1), and 2 miRNAs (miR-146a-5p and miR-92a-3p) were selected on the basis of combined adjusted P values <.1 and differential expression in previous MCL studies.18,29 Four additional miRNAs (miR-29a, miR-29b, miR-29c, and miR-20b), also known from the literature30,31 but not differentially expressed in our microarray analysis, were also chosen for qRT-PCR analysis in MCL2. Three miRNAs were selected as reference miRNAs on the basis of their low variance of expression across all microarrays (supplemental Figure 1).
With regard to CSS, 7 of the miRNAs selected for qRT-PCR analysis (miR-4417, miR-18b-5p, miR-3687, miR-378d, miR-185-5p, miR-144-3p, and miR-92a-3p) were differentially expressed, thus confirming the microarray findings in the MCL2 cohort (Table 1). Log fold changes tended to be higher in the qRT-PCR analysis compared to the microarray, and the adjusted P values were correspondingly lower by qRT-PCR than by microarray.
High expression of miR-18b predicts poor prognosis in MCL
Previous studies have identified various miRNAs that predict outcome in heterogeneous cohorts of MCL patients16,17 ; however, the prognostic value of miRNA profiles in similarly treated cohorts of MCL patients remains to be demonstrated. To evaluate the prognostic performance of the differentially expressed miRNAs, we performed minimal-redundancy-maximal-relevance analysis and a leave-one-out cross-validation using linear discriminant analysis. The maximum enhancement of the prognostic accuracy of the MIPI-B was achieved by miR-18b expression (supplemental Figure 2); in addition, a univariate analysis revealed that miR-18b overexpression was significantly correlated with poor survival in MCL2 (P < .001, log-rank) (supplemental Figure 3).
Validation in the independent MCL3 cohort
To validate findings from the MCL2 cohort, we analyzed the confirmed differentially expressed miRNAs in the independent but similarly treated MCL3 cohort by qRT-PCR. Here, we were able to validate miR-18b, miR-378d, and miR-92a as being significantly overexpressed (adjusted P < .05) in patients who died of MCL (Table 1). Both miR-18b and miR-92a have previously been associated with aggressive subtypes of MCL, whereas miR-378d is a novel prognostic factor for survival in MCL.
MIPI-B-miR: addition of miR-18b expression improves the prognostic value of the MIPI-B
Several studies, including the Nordic MCL2 trial,7 have confirmed the MIPI-B score as the best performing prognosticator of MCL outcome. To assess the prognostic value of miRNA expression, we incorporated the MIPI-B components with expression data of miR-18b. Based on the MCL2 screening cohort, we developed a novel prognostic index using linear discriminant analysis of miR-18b expression levels and the MIPI-B value to assign patients to high, intermediate, or low risk. This new prognostic index, the MIPI-B-miR, was defined as: , where
, where  is the MIPI-B value, x is the log-fold-change of miR-18b, and c is a regression parameter (c = 0.58317). Low risk was defined as
is the MIPI-B value, x is the log-fold-change of miR-18b, and c is a regression parameter (c = 0.58317). Low risk was defined as  , high risk was defined as
, high risk was defined as  , and intermediate risk is implied when MIPI-B-miR values fall between these thresholds (see supplemental text).
, and intermediate risk is implied when MIPI-B-miR values fall between these thresholds (see supplemental text).
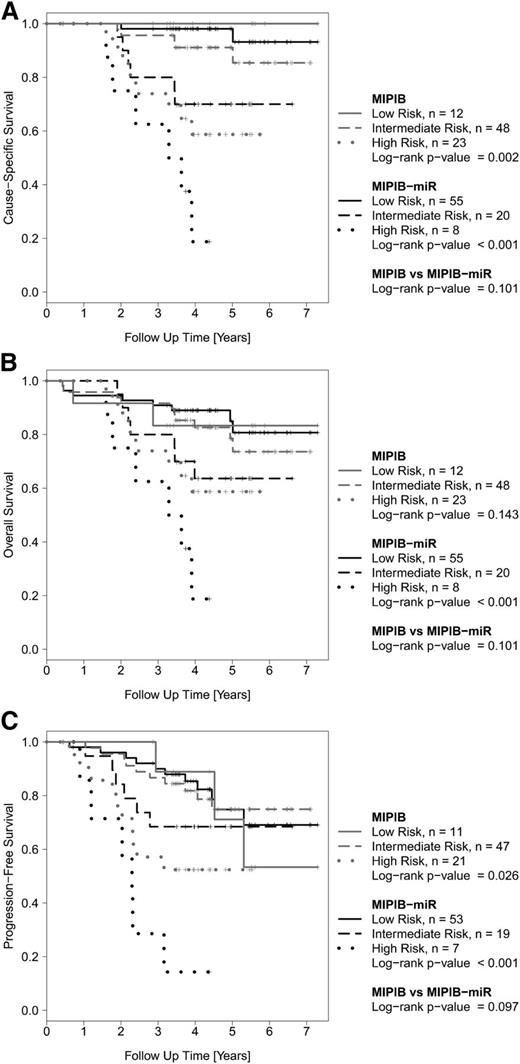
The MIPI-B-miR had a significantly better separation of patients in low, intermediate, and high risk groups compared to the standard MIPI-B regarding OS and PFS, and was borderline significant with regard to CSS (Figure 3). In particular, the MIPI-B-miR reduced the number of high-risk patients by two-thirds and identified a group of patients with an exceedingly poor outcome compared to that identified by the MIPI-B high-risk index.
Outcome of MCL patients in the screening cohort (MCL2) according to the MIPI-B and MIPI-B-miRNA prognostic models. The MIPI-B prognosticator (grey) divides the cohort into 3 risk groups according to established cutoff values. The MIPI-B-miR prognosticator (black), which also includes the qRT-PCR expression level of miR-18b, also divides the cohort into 3 risk groups. Kaplan-Meier estimates of CSS (A), OS (B), and PFS (C).
Outcome of MCL patients in the screening cohort (MCL2) according to the MIPI-B and MIPI-B-miRNA prognostic models. The MIPI-B prognosticator (grey) divides the cohort into 3 risk groups according to established cutoff values. The MIPI-B-miR prognosticator (black), which also includes the qRT-PCR expression level of miR-18b, also divides the cohort into 3 risk groups. Kaplan-Meier estimates of CSS (A), OS (B), and PFS (C).
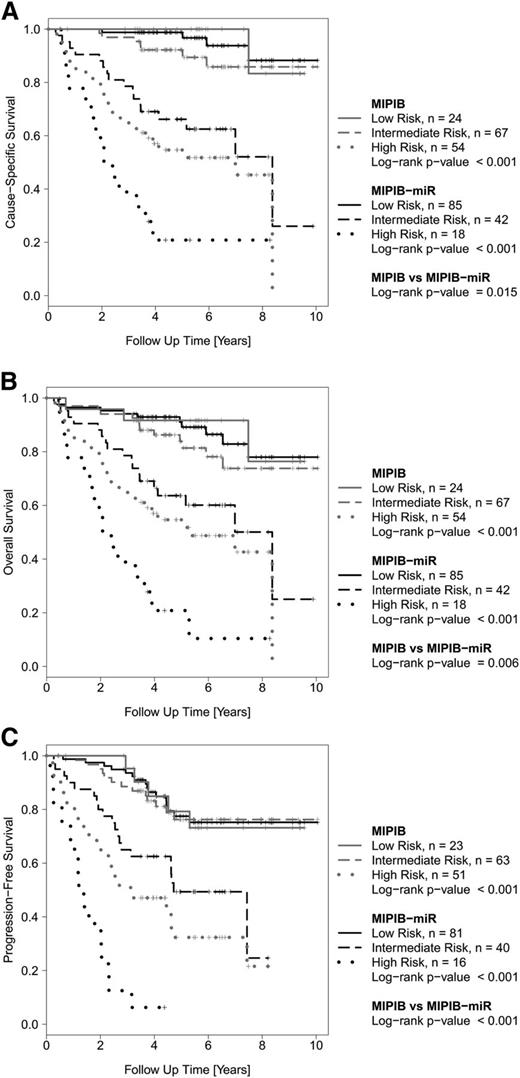
To confirm these findings, we blindly tested the MIPI-B-miR prognosticator on the independent MCL3 validation cohort. The MIPI-B-miR also improved the separation of risk groups in the validation cohort regarding CSS, OS, and PFS (Figure 4). For example, the MIPI-B-miR achieved significant prognostic value for OS (P < .001), whereas the MIPI-B did not (P = .143). Compared to the MIPI-B, the MIPI-B-miR showed a trend of improved identification of high-risk patients (CSS: P = .101; OS: P = .101; and PFS: P = .097), despite the shorter observation time in this cohort.
Outcome of MCL patients in the validation cohort (MCL3) according to the MIPI-B and MIPI-B-miR prognostic models. Kaplan-Meier estimates of CSS (A), OS (B), and PFS (C).
Outcome of MCL patients in the validation cohort (MCL3) according to the MIPI-B and MIPI-B-miR prognostic models. Kaplan-Meier estimates of CSS (A), OS (B), and PFS (C).
Based on the improved prognostication by the MIPI-B-miR, we next applied this index to the entire patient material (MCL2 and MCL3). In the combined analysis, the MIPI-B-miR was superior to the standard MIPI-B at identifying high-risk patients with respect to CSS, OS, and PFS (Figure 5 and supplemental Figure 4). In the subset of patients with a known MRD marker, the MIPI-B-miR predicted a high risk of molecular relapse (P < .001), whereas the MIPI-B did not (P = .322; supplemental Figure 5). Importantly, the MIPI-B-miR was still significantly superior to the MIPI-B at predicting CSS (P = .034), OS (P = .034), and PFS (P = .006) when patients receiving preemptive rituximab were excluded from the analysis of the combined cohort (MCL2 and MCL3) (supplemental Figure 6). Finally, a multivariate analysis of the combined MCL2 and MCL3 cohorts confirmed miR-18b as an independent prognostic factor for OS (supplemental Table 3).
Outcome of MCL patients in the combined similarly treated cohorts (MCL2 and MCL3) according to the MIPI-B and MIPI-B-miR prognostic models. Kaplan-Meier estimates of CSS (A), OS (B), and PFS (C).
Outcome of MCL patients in the combined similarly treated cohorts (MCL2 and MCL3) according to the MIPI-B and MIPI-B-miR prognostic models. Kaplan-Meier estimates of CSS (A), OS (B), and PFS (C).
MiR-18b function
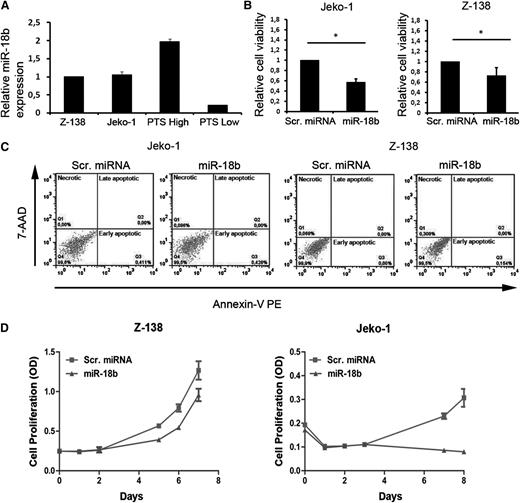
To investigate the possible functional consequences of miR-18b overexpression, 2 MCL cell lines, Z-138 and Jeko-1, were selected due to their high transfection efficiency. Both cell lines express miR-18b at intermediate levels compared to primary MCLs expressing high or low levels of miR-18b (Figure 6A). Z-138 and Jeko-1 cells transfected with miR-18b mimic display significantly lower viability scores compared to cells transfected with scrambled miRNA as measured by XTT assay (P = .05 and P = .005, respectively) (Figure 6B). The low cell viability observed on miR-18b overexpression was not associated with the induction of apoptosis, because no increase in the early apoptotic cell population was detected when miR-18b-transfected cells were compared to scrambled miRNA–transfected cells (Figure 6C). However, when comparing the proliferation rate between cells transfected with miR-18b or scrambled miRNA, miR-18b was found to markedly inhibit the proliferative ability of both Z-138 and Jeko-1 cells over an 8-day test period, although this was most pronounced in Jeko-1 cells (Figure 6D). Overexpression of miR-18b in Jeko-1 and Z-138 cells did not alter their cell cycle profile when compared to scrambled miRNA–transfected cells (data not shown), indicating that miR-18b likely inhibits cell cycle progression at multiple points.
MiR-18b inhibits cell proliferation in Jeko-1 and Z-138 cells lines. (A) Relative expression levels of miR-18b in Z-138 and Jeko-1 compared with MCL patient samples expressing high (PTS High) or low (PTS Low) miR-18b. Results are expressed as means ± standard deviation (SD). (B) Cells were transfected with 1 μM scrambled (Scr.) miRNA control or miR-18b mimic for 6 days, after which cell numbers were determined by XTT assay. Results are expressed as means ± SD. Statistical analyses were performed using the Student t test. *P ≤ .05. (C) Apoptosis was studied by flow cytometry in Annexin V-PE/7 AAD–stained cells 24 hours after transfection with Scr. miRNA control or miR-18b mimic. (D) Growth curves for cells transfected with Z-138 and Jeko-1. Cell number was determined 0 to 8 days after transfection using the XTT assay. Results are expressed as means ± SD. OD, optical density.
MiR-18b inhibits cell proliferation in Jeko-1 and Z-138 cells lines. (A) Relative expression levels of miR-18b in Z-138 and Jeko-1 compared with MCL patient samples expressing high (PTS High) or low (PTS Low) miR-18b. Results are expressed as means ± standard deviation (SD). (B) Cells were transfected with 1 μM scrambled (Scr.) miRNA control or miR-18b mimic for 6 days, after which cell numbers were determined by XTT assay. Results are expressed as means ± SD. Statistical analyses were performed using the Student t test. *P ≤ .05. (C) Apoptosis was studied by flow cytometry in Annexin V-PE/7 AAD–stained cells 24 hours after transfection with Scr. miRNA control or miR-18b mimic. (D) Growth curves for cells transfected with Z-138 and Jeko-1. Cell number was determined 0 to 8 days after transfection using the XTT assay. Results are expressed as means ± SD. OD, optical density.
Discussion
This study of 168 patients provides the most comprehensive study of miRNA expression profiles in MCL to date. Moreover, it is the first study to prospectively follow and analyze miRNA expression in a large homogeneously treated cohort of MCL patients. The detailed trial data also enabled us to study CSS, excluding treatment-related deaths or deaths from other causes. By analyzing the expression of 1896 human miRNAs by microarray in patients from the MCL2 cohort, we identified 17 miRNAs that were significantly upregulated or downregulated in patients who died of MCL. qRT-PCR analysis confirmed that 7 of these miRNAs were upregulated. Subsequent validation in the independent MCL3 cohort identified an aggressive MCL subtype, biologically characterized by aberrant overexpression of miR-18b, miR-92a, and miR-378d. These miRNAs, and miR-18b in particular, signify MCL patients who are incurable by the current standard therapy of rituximab (R)–maxi–CHOP (cyclophosphamide, hydroxy doxorubicin, vincristine, and prednisone) alternating with R-high-dose cytarabine (6 cycles total), followed by high-dose BEAM/C (bis-chloroethylnitrosourea, etoposide, cytarabine, and melphalan or cyclophosphamide) and ASCT.
The prognostic impact of miRNAs has previously been studied in MCL, but mainly in smaller heterogeneous cohorts treated with different regimens.16,17 These studies have suggested a number of prognostic miRNAs, including miR-18b29 and the entire miR-17∼92a cluster. However, no other study identified miR-92a as a key component. Here, we confirm that these miRNAs are valid prognostic markers in our large cohort of homogeneously treated MCL patients. Moreover, we observed that other, previously published prognostic miRNAs (miR-146a, miR-20b, miR-29a, and miR-29c)29-31 displayed trends in our data that agreed with earlier findings but were not statistically significant in this study.
From a genomic perspective, miR-18b and miR-92a are located in 2 paralogue clusters (the miR-106a∼363 cluster on chromosome X and the miR-17∼92a cluster on chromosome 13, respectively) with shared evolutionary origin.32 In particular, overexpression of the miR-17∼92a cluster has been widely studied and implicated in both benign and malignant lymphoproliferative diseases,33 whereas the functions of the miRNAs in the miR-106a∼363 cluster, which share numerous seed sequences with the 17∼92a cluster miRNAs, are less well explored.34 Overexpression of miR-378d has been associated with poor prognosis in acute myeloid leukemia35 and with brain metastasis in non-small cell lung cancer36 but, to our knowledge, has not been implicated in lymphoid malignancies.
Although aberrantly expressed miRNAs have been suggested as prognostic markers for survival in MCL, no study has yet identified miRNAs that could improve on the state-of-the-art prognosticator, the MIPI-B.16,17 In the current study of 2 similar MCL trial cohorts, we found that the miR-18b expression level significantly adds prognostic information to the MIPI-B approach. By generating a new prognosticator, the MIPI-B-miR, we were able to predict survival of MCL patients after high-dose immunochemotherapy and ASCT and improve the gold standard prognosticator, the MIPI-B. Because the MIPI-B-miR was validated in a similarly treated cohort and was also found to be significant in the whole test group, we believe that this prognosticator may also have prognostic power in other cohorts of younger MCL patients treated by current standards (rituximab plus chemotherapy and ASCT). In addition, the MIPI-B-miR was superior to the MIPI-B in predicting molecular relapse. Because exclusion of cases treated with preemptive rituximab did not change the overall picture, the MIPI-B-miR may also be valid in settings without MRD surveillance and preemptive therapy. Future investigations using the MIPI-B-miR will be required to demonstrate its validity in differently treated and elderly cohorts of MCL patients. In this study, quantification of miRNAs was performed by standard primer-based qRT-PCR on RNA extracted from FFPE preserved MCL specimens, which are easily storable and accessible, emphasizing the feasibility and the robustness of the method.
Beyond their value as prognostic markers, aberrantly expressed miRNAs may be therapeutic targets in patients,37 potentially enabling new therapies in the poor-prognosis group. It is unfortunate that the direct functions of these upregulated miRNAs in MCL are largely unknown. In particular, divergent roles of miR-18b in carcinogenesis have been reported and complicate therapeutic approaches. For example, low expression of miR-18b has been shown to predict melanoma progression and short survival,38 whereas in hepatocellular carcinoma, high miR-18b expression correlates with low survival rate.39 The dual role of miR-18b in cancer is not surprising, considering that different functions have been previously attributed to individual miRNAs in different cellular settings.12 Interestingly, high expression of miR-18b is characteristic of human embryonic stem cells.40
We have investigated miR-18b function in 2 MCL cell lines and observed that miR-18b overexpression significantly downregulated MCL cell proliferation. This observation supports that miR-18b adds prognostic information to the MIPI-B by identifying an additional group of patients with poor prognosis, not identified by the MIPI-B, which includes the proliferation marker Ki-67. The regimens used in the Nordic MCL2 and MCL3 trials comprise several cell cycle–specific drugs, including cytarabine, which target only proliferating cells. Our in vitro studies clearly show an association between high miR-18b expression and low proliferation rate. We therefore suggest that high miR-18b expression may render MCL cells resistant to the MCL2/MCL3 therapeutic regimens, leading to poor prognosis. Inhibition of cell proliferation by miRNAs has previously been shown to play a pivotal role in chemoresistance; for example, in osteosarcoma and colon cancer cells, in which both miR-140 and miR-215 cause chemoresistance by reducing the proliferation rate.41,42 Because high expression of miR-18b is also typical of undifferentiated cells and embryonal carcinoma cells,40,43 it is tempting to speculate whether miR-18b expression can induce stemness in MCL, which is a subject for further studies.
The microarray data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE66299).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Professor Finn Cilius Nielsen and laboratory technicians at the Center for Genomic Medicine, Rigshospitalet, for providing essential equipment.
This work was supported by grants from The Danish Cancer Society (S.H., U.R., K.G., J.B.C., R.Z., and C.H.G.); and by Rigshospitalet’s Research Foundation (K.G. and C.H.G.); the Novo Nordisk Foundation (K.G., J.B.C., and C.H.G.); the Danish Council for Strategic Research, Center for non-coding RNA in Technology and Health (C.G. and C.T.W.); and the Danish Medical Research Council (R.Z.).
Authorship
Contribution: S.H., U.R., R.Z., J.B.C., C.H.G., and K.G. conceived and designed the experiments; S.H., U.R., R.Z., L.B.P., and A.P. performed the experiments; S.H., U.R., R.Z., J.B.C., C.G., C.T.W., P.B., and K.G. analyzed the data; S.E., A.K., M.J., A.L., R.R., M.E., C.S., M.-L.K.-L., J.D., E.C.-L., P.B., C.H.G., and K.G. contributed with patient material/reagents/materials/analysis tools; and S.H., U.R., R.Z., J.B.C., C.G., C.T.W., C.H.G., and K.G. wrote the paper. All authors read and approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Kirsten Grønbæk, Department of Hematology, L4042, Rigshospitalet, Blegdamsvej 9, DK-2100 Copenhagen Ø, Denmark; e-mail: kirsten.groenbaek@regionh.dk.
References
Author notes
S.H. and U.R. contributed equally to this study.
C.H.G. and K.G. contributed equally to this study.