In this issue of Blood, Micol et al report the discovery of mutations in the additional sex combs-like 2 (ASXL2) gene in about one-quarter of acute myeloid leukemia (AML) patients carrying the t(8;21) translocation.1
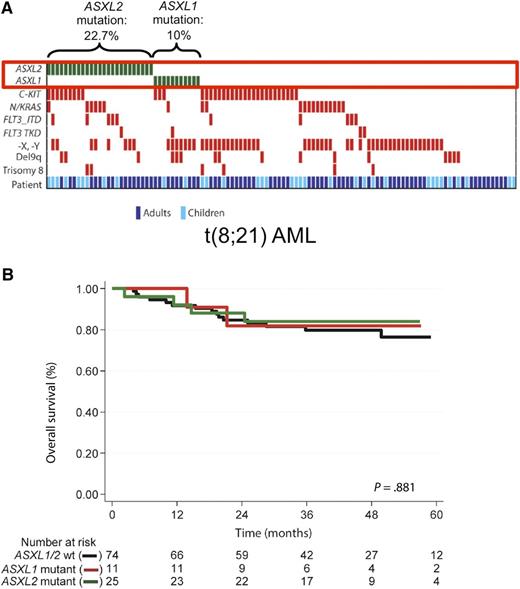
ASXL gene mutations in t(8;21) AML. (A) ASXL2 is the second-most commonly mutated gene in AML with t(8;21). The heatmap visualizes the spectrum of recurrent gene mutations in 110 adult and pediatric patients with AML and t(8;21). ASXL2 and ASXL1 mutations were mutually exclusive. The figure has been adapted from Figure 1 in the article by Micol et al that begins on page 1445. (B) ASXL2 gene mutations do not affect overall survival in t(8;21) AML. The Kaplan-Meier curves depict overall survival of patients with mutated ASXL1, mutated ASXL2, or wild-type ASXL1/ASXL2. No difference between the 3 groups was observed. The figure has been adapted from supplemental Figure 4 in the article by Micol et al that begins on page 1445.
ASXL gene mutations in t(8;21) AML. (A) ASXL2 is the second-most commonly mutated gene in AML with t(8;21). The heatmap visualizes the spectrum of recurrent gene mutations in 110 adult and pediatric patients with AML and t(8;21). ASXL2 and ASXL1 mutations were mutually exclusive. The figure has been adapted from Figure 1 in the article by Micol et al that begins on page 1445. (B) ASXL2 gene mutations do not affect overall survival in t(8;21) AML. The Kaplan-Meier curves depict overall survival of patients with mutated ASXL1, mutated ASXL2, or wild-type ASXL1/ASXL2. No difference between the 3 groups was observed. The figure has been adapted from supplemental Figure 4 in the article by Micol et al that begins on page 1445.
Patients with this chromosomal translocation are classified as having “favorable-risk” disease, but <60% of them are actually cured by current treatment approaches.2 Activating mutations in the KIT gene occur in approximately one-third of patients with t(8;21) and identify a subgroup with adverse outcomes. However, even among patients with wild-type KIT, the relapse rate exceeds 30%. Expanding our knowledge about cooperating genetic events in AML with t(8;21) may uncover additional prognostic markers and reveal novel therapeutic targets.
Based on this rationale, Micol et al performed whole-exome sequencing of 3 patients with t(8;21) AML, and identified 2 somatic mutations in the ASXL2 gene. In a validation cohort of 110 adult and pediatric patients with t(8;21), ASXL2 mutations were present in 23%, making ASXL2 the second-most commonly mutated gene in this cytogenetic subgroup, behind KIT (see figure). ASXL2 mutations have previously been reported, although at a much lower frequency, in various solid tumors including cancers of the bladder, prostate, endometrium, pancreas, and breast. Recently, Huether and colleagues studied 1000 pediatric cancer genomes and found ASXL2 mutations in 7 patients, including 6 with AML and balanced chromosomal rearrangements affecting the core binding factor (CBF) genes.3 Micol and colleagues now extend this initial report by demonstrating that ASXL2 mutations are restricted to patients with the t(8;21) translocation, which leads to a RUNX1-RUNX1T1 gene fusion. In contrast, ASXL2 mutations were absent in patients with RUNX1 point mutations or in patients with inversion inv(16), causing rearrangement of the RUNX1 heterodimerization partner CBF β (CBFB). ASXL1 mutations were found in 10% of t(8;21) patients, and were mutually exclusive with ASXL2 mutations.
Micol et al also investigated the prognostic relevance of ASXL gene mutations in t(8;21) AML. They observed no difference in overall survival between ASXL1-mutated, ASXL2-mutated, and wild-type patients (see figure). Cumulative incidence of relapse appeared to be higher in patients with ASXL1 or ASXL2 mutations, but this difference did not achieve statistical significance. These results are consistent with a previous study reporting that 11.5% of adult t(8;21) AML patients carry ASXL1 mutations, and that ASXL1-mutated patients have significantly shorter event-free survival, but similar overall survival compared with those with wild-type ASXL1.4 Of note, we and others have found that ASXL1 gene mutations associate with inferior outcomes in patients with cytogenetically normal AML.5 Several caveats need to be taken into account when interpreting the results in t(8;21) patients. In both studies,1,4 patient numbers were relatively small. Therefore, it was not possible to perform multivariable analyses, or to assess the relevance of ASXL1 and ASXL2 mutations in the patient subgroups with mutated and wild-type KIT. Furthermore, Micol et al jointly analyzed adult and pediatric patients treated on different protocols. Thus, additional studies in larger, well-defined cohorts are needed to clarify the prognostic relevance of ASXL gene mutations in t(8;21) AML. These results illustrate a challenge in translational AML research: due to the increasing number of known recurrent gene mutations, we can delineate more and more patient subsets characterized by specific combinations of genetic alterations. However, reliably establishing the clinical significance of such small subgroups requires large patient cohorts, which often will only be available through collaborations between different study groups.
What do the results of Micol et al teach us about the pathogenesis of AML? ASXL2 belongs to a human gene family with 3 members (ASXL1, ASXL2, and ASXL3) that are homologs of the Drosophila additional sex combs (Asx) gene. Mutations in ASXL1 were initially described in 2009 in patents with myelodysplastic syndromes or chronic myelomonocytic leukemia,6 and also occur in AML and myeloproliferative neoplasms. ASXL1 knockout in the murine hematopoietic system causes myelodysplasia-like changes. This phenotype is associated with deregulation of the polycomb-repressive complex PRC2 and global reduction of histone H3 lysine 27 (H3K27) trimethylation, a chromatin modification associated with gene silencing.7 Similar to ASXL1, the ASXL2 protein is involved in regulation of the PRC2 complex. ASXL2 also interacts with BAP1, a protein involved in cell-cycle regulation, and with nuclear hormone receptors such as the retinoic acid receptor and peroxisome proliferator-activated receptor-γ.8 However, the function of ASXL2 in normal and malignant hematopoiesis is largely unknown. Silencing of ASXL2 in mice results in partial embryonic lethality, and shortened lifespan of adult animals along with skeletal and cardiac abnormalities.8 Hematological abnormalities have not been described in this model, and a tissue-specific knockout of ASXL2 in the hematopoietic system has not yet been reported. The report by Micol and colleagues should therefore prompt further research into the role of ASXL2 in hematopoiesis.
In this context, it is an interesting observation that ASXL gene mutations are strongly associated with alterations of the RUNX1 gene. In line with the report by Micol et al, Pratcorona and coworkers found ASXL1 mutations in 8% of t(8;21) patients, but not in patients with inv(16).9 Another study in cytogenetically normal AML showed that 35% of patients with mutated RUNX1 carried an ASXL1 mutation, compared with only 6% of RUNX1 wild-type patients.10 These data support a functional cooperation between RUNX1 alterations, which are also known to cause epigenetic deregulation, and ASXL gene mutations. Because ASXL1 and ASXL2 mutations were mutually exclusive in t(8;21) AML, it is likely that both gene mutations affect similar cellular pathways. On the other hand, ASXL2 mutations appear to be restricted to patients with the RUNX1-RUNX1T1 gene fusion, whereas ASXL1 mutations are also found in patients with RUNX1 point mutations, indicating that the effects of mutations in the 2 genes are not identical. In summary, the results by Micol and others strongly suggest that there is a functional connection between RUNX1, a central regulator of normal and malignant hematopoiesis, and ASXL gene mutations. Uncovering this link is an exciting topic for future research that will not only improve our insights into the role of the ASXL gene family in leukemogenesis, but may also lead to specific therapeutic approaches targeting the ASXL-RUNX1 interaction.
Conflict-of-interest disclosure: The author declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal