In this issue of Blood, Moorman et al show that most good-risk patients can now be classified robustly by integrating the information from prevalent copy-number alterations (CNAs) in relevant combinations and classical cytogenetic risk factors in acute lymphoblastic leukemia (ALL).1
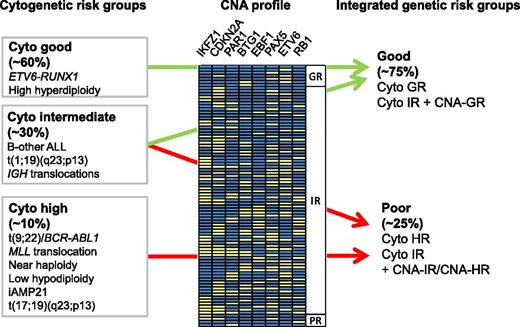
Integration of cytogenetic risk groups and CNA profile information. CNAs are depicted as yellow boxes. The profiles are ordered according to their associated risk (GR, good risk; IR, intermediate risk; PR, poor risk). For CNA data related to this figure, see supplemental Table 3 in the article by Moorman et al, which begins on page 1434. Figure kindly provided by A.V. Moorman, University of Newcastle.
Integration of cytogenetic risk groups and CNA profile information. CNAs are depicted as yellow boxes. The profiles are ordered according to their associated risk (GR, good risk; IR, intermediate risk; PR, poor risk). For CNA data related to this figure, see supplemental Table 3 in the article by Moorman et al, which begins on page 1434. Figure kindly provided by A.V. Moorman, University of Newcastle.
ALL is a complex genetic disease that results from the combination of lesions in genes involved in the regulation of hematopoiesis, lymphoid differentiation, cell cycle, and proliferation. High-resolution genetic profiling of ALL reveals an increasing number of underlying mutations that modify the function of transcription factors, epigenetic regulators, or components of signaling pathways, among others.2 For decades, cytogeneticists have been aware of specific chromosomal translocations, some of which have been established as reliable risk factors (see figure). However, these genetic risk factors identify only some of the patients at risk. Most study groups therefore also include the assessment of minimal residual disease (MRD) during induction chemotherapy for risk stratification, a powerful approach that was pioneered by European study groups.3,4 The challenge is now to devise new approaches to translate the rapidly expanding knowledge of ALL genomics to the clinic in order to define better prognostic markers and specific druggable targets already at diagnosis. The most common submicroscopic genetic lesions include CNAs or sequence mutations of hematopoietic transcription factors, including PAX5, IKZF1, and EBF1; gene rearrangements leading to overexpression of the cytokine receptor component CRLF2; mutations in JAK1 and JAK2 kinases; and deletions of the CDKN2A/B loci encoding the INK4/ARF tumor suppressor genes, to name representative examples.2 Several of these lesions have been associated with less favorable outcome (such as CDKN2A/CDKN2B and IKFZ1 deletions5,6 and P2RY8-CRLF2 fusion7 ), although a clear consensus for their clinical application as prognostic markers is lacking. Evidence for prognostic significance of recurrent constellations of such genetic lesions has emerged in the field. One of the first examples for such a relevant combination is the strong association of IKFZ1 deletions with BCR-ABL1–positive ALL.8 Thus, the integration of multiple parameters to identify recurrent patterns may lead to better options for clinical management.
In the present study,1 Moorman et al took advantage of multiplex ligation-dependent probe amplification to detect the most frequent CNAs retrospectively in >1600 patients treated in the ALL97/99 (n = 864) and UKALL2003 (n = 742) trials in order to establish constellations of CNAs that would be associated with outcome. Using ALL97/99 as a test cohort, they determined the prognostic relevance of different CNA combinations and integrated the resulting 3 CNA risk groups with 3 cytogenetic risk groups to define 2 genetic risk groups. One strength of the study was that the classification was then validated using an independent cohort of patients treated in the UKALL2003 trial. Detection of CNA constellations that were associated with good risk enabled the reclassification of half of the patients with intermediate risk by cytogenetics into a group with favorable outcome (see figure). In fact, good-risk patients with low MRD after induction chemotherapy had an overall survival of 99%. The immediate potential of this new approach for genetic classification is to identify favorable-risk patients who may qualify for treatment de-escalation. This will provide better tools for early interventions in clinical trials in an attempt to reduce toxicity.
This study illustrates at a very practical level the need for a systems-oriented analytical approach to personalize ALL treatment. On this path, we need to ingrate high-resolution genetic profiling with all other available layers of diagnostic and clinical information. For example, a subset of patients in the genetic good-risk group in this study remained MRD positive, which was associated with significantly more relapses but still an overall survival >90%, indicating that salvage was possible in most cases. The drivers of this de novo drug-resistance phenotype remain to be discovered and limit the currently available genetic classification. Similarly, the relevant genetic constellations of most high-risk patients who do not carry a prognostic cytogenetic marker (the so-called B-others) need systematic exploration. Of obvious importance among the B-other group is BCR-ABL1–like leukemia with a gene expression profile that resembles BCR-ABL1 ALL.9 Next-generation sequencing of representative cases revealed a wide range of genetic alterations leading to activation of cytokine-receptor and kinase signaling, involving ABL1, ABL2, EPOR, IL7R, JAK2, CSF1R, and PDGFRB, for example.10 Informative constellations also emerge here, such as the association with IKFZ1 deletions, CDKN2A/B deletions, or P2RY8-CRLF2 fusions in patients with poor risk based on MRD.
In conclusion, this study provides a widely applicable strategy to improve the definition of good-risk patients in ALL, which could also be implemented in countries with more limited access to high-end genomic technology. It represents an important step toward a more integrated use of biomedical data to guide ALL therapy. We can expect the rapid discovery of additional relevant ALL molecular phenotypes and the translation of this knowledge in interventional studies for the benefit of our patients.
Conflict-of-interest disclosure: The author declares no competing financial interests.