Key Points
The miR-142-3p double mutant zebrafish displayed aberrant neutrophil hypermaturation and homeostasis in myelopoiesis.
Abnormal activation of IFN-γ signaling mediated the impaired neutrophil development in miR-142-3p–deficient zebrafish.
Abstract
Neutrophils play critical roles in vertebrate innate immune responses. As an emerging regulator in normal myelopoiesis, the precise roles of microRNA in the development of neutrophils have yet to be clarified. Using zinc-finger nucleases, we have successfully generated heritable mutations in miR-142a and miR-142b and showed that hematopoietic-specific miR-142-3p is completely deleted in miR-142 double mutant zebrafish. The lack of miR-142-3p resulted in aberrant reduction and hypermaturation of neutrophils in definitive myelopoiesis, as well as impaired inflammatory migration of neutrophils in the fetal stage. Furthermore, the adult myelopoiesis in the miR-142-3p–deficient zebrafish was also affected, producing irregular hypermature neutrophils with increased cell size and a decreased nucleocytoplasmic ratio. Additionally, miR-142-3p–deficient zebrafish are expected to develop a chronic failure of myelopoiesis with age. Transcriptome analysis showed an aberrant activation of the interferon γ (IFN-γ) signaling pathway in myelomonocytes after miR-142-3p deletion. We found that the reduced number and hypermaturation of neutrophils caused by loss of miR-142-3p was mainly mediated by the abnormally activated IFN-γ signaling, especially the upregulation of stat1a and irf1b. Taken together, we uncovered a novel role of miR-142-3p in maintaining normal neutrophil development and maturation.
Introduction
The neutrophil, as a pivotal component of the innate immune system, is generated through normal myelopoiesis. Myelopoiesis is a tightly regulated developmental process in which myeloid progenitors become mature myeloid cells through hematopoiesis. Studies have revealed the necessary role of several transcription factors in normal vertebrate myelopoiesis, including Pu.1, Cebpa, Cebpe, Irf8, and Gfi1,1-5 but the orchestrated regulatory program in normal myelopoiesis remains unclear.
Emerging evidence has shown that microRNAs (miRNAs), ∼22-nucleotide (nt) posttranscriptionally regulatory RNAs, are widely involved in normal myelopoiesis. In murine granulocyte-macrophage progenitors, the deletion of Dicer1, an essential endoribonuclease in the process of pre-miRNAs, caused myeloid dysplasia and depleted macrophages.6 The myeloid-specific miR-223 fine-tunes the differentiation of granulocytes.7 Several other miRNAs, including miR-146a, miR-21, and miR-196b, also play an important role downstream of key transcription factors in normal myelopoiesis, like PU.1 and Gfi1.8,9
miR-142-3p was first described as a hematopoietic-specific miRNA in mice.10 Studies have elucidated multiple regulatory roles of miR-142-3p in hematopoietic development. For example, knockdown of miR-142-3p with a morpholino in Xenopus impairs the specification of a definitive hemangioblast.11 The dysregulated expression of miR-142-3p could affect the proliferation of the CD25+ CD4+ T-cell response to stimulation in vitro.12 In addition, the forced expression of miR-142-3p improves myeloid differentiation in leukemia cell lines.13 A controversial role of miR-142-3p in macrophage differentiation was also reported for cancer-induced myelopoiesis.14 However, all of these studies were based on overexpression or knockdown experiments. The functional study of this miRNA in knockout animal models remains limited, except in the case of recent study focusing on a homeostatic defect of CD4+ dendritic cells in miR-142-3p–deficient mice.15 Hence, detailed functions of miR-142-3p in vivo require clarification.
Zebrafish has been developed as a good complementary model animal for studying myelopoiesis. Hematopoiesis in zebrafish was very similar to that in mammals, in which definitive hematopoiesis rapidly replaces the primitive hematopoiesis and gives rise to many types of blood cells.16 Most of the myeloid cell types in mammals have also been identified in zebrafish,17-21 including neutrophils, eosinophils, mast cells, monocytes/macrophages, and dendritic cells. The transparent embryos, external development, and feasible transgenic lines that specifically label myeloid cells in zebrafish facilitate the elucidation of myeloid-specific developmental programs in vivo.
Zinc-finger nucleases (ZFNs), a well-established tool for inducing heritable mutation within a target gene locus, are applicable in different species.22-25 Here, we successfully disrupted the miR-142a and miR-142b genes in zebrafish using ZFN-mediated gene targeting with the oligomerized pool engineering (OPEN) method and context-dependent assembly (CoDA) approach, respectively.26-28 We showed that deletion of miR-142-3p in the miR-142a/b double mutant zebrafish led to aberrant myelopoiesis, which produced abnormal neutrophils, and further provided the involved mechanisms in this study.
Methods
Zebrafish lines and maintenance
Wild-type Tuebingen zebrafish and transgenic zebrafish lines, including Tg(zlyz:eGFP) and Tg(cmyb:eGFP), were raised and maintained under standard conditions. The zebrafish experiments were approved by the Institutional Review Board of the Institute of Health Sciences, Shanghai Institutes of Biological Sciences, Chinese Academy of Sciences (Shanghai, China).
Selection and construction of ZFNs
Generation of zebrafish miR-142a and miR-142b mutant lines
Capped ZFN-encoding RNAs specific for miR-142a or miR-142b were synthesized (mMESSAGE mMACHINE T7 Ultra kit; Ambion, Austin, TX) and then capped with a poly(A) tail (Poly[A] Tailing Kit; Ambion) as described previously.29 For efficient induction of mutagenesis in miR-142a, ∼100 pg of 142aZFN-coding RNAs were injected into 1-cell stage zebrafish embryos, which resulted in an appropriate 80% rate of deformity at 24 hours postfertilization (hpf). For mutagenesis in miR-142b, the injection dose of 142bZFN-coding RNAs was ∼200 pg, and the rate of deformity was an appropriate 70%. The surviving embryos after injection with ZFN-coding RNAs were raised to adulthood and screened for founders with germ-line mutations in the miR-142a or miR-142b locus, respectively.
Synthesis of morpholinos, miRNA duplex, and mRNA for microinjection
Morpholinos (Gene Tools) were synthesized in Gene Tools and injected into 1-cell stage embryos. The sequence and dosage for each morpholino are detailed in supplemental Table 3. The miR-142-3p duplex and its 2-bp mismatch control were synthesized at GenePharma (Shanghai, China). Messenger RNAs (mRNAs) for wild-type or mutant pre-miR-142a/b were transcribed from linearized plasmid templates using the mMESSAGE mMACHINE Kit (Ambion).
Whole-mount in situ hybridization and Sudan black staining
Digoxigenin (DIG)–labeled RNA probes were transcribed from linearized DNA templates with SP6 or T7 RNA polymerase (Ambion). Whole-mount in situ hybridization (WISH) for the detection of hematopoietic markers were performed at 66°C, as described previously.30,31 WISH with double DIG-labeled lock nuclear acid (LNA) probe (Exiqon) for dre-miR-142a-3p or dre-miR-142a-5p was performed at 57.5°C. The Sudan black staining of neutrophils was performed as described previously.32
Flow cytometry and Wright-Giemsa staining
Transgenic Tg(zlyz:eGFP) and Tg(cmyb:eGFP) zebrafish embryos at 3 days postfertilization (dpf) were rinsed in phosphate buffered saline, disassociated in 0.25% trypsin/EDTA (Invitrogen) for 35 minutes at 37°C, and filtered through a 40-μm cell strainer (BD Falcon). Enhanced green fluorescent protein (EGFP)–positive cells were sorted with a MoFlo FACS sorter (Dako Cytomation). See supplemental Methods for the sorting of zebrafish whole kidney marrow (WKM).
The Wright-Giemsa staining of sorted cells was performed with the Wright-Giemsa staining kit (Baso Diagnostic, Inc.) according to the manufacturer’s instructions.
mRNA expression profiling
The myelomonocyte fraction and WKM (without erythrocytes) were sorted from wild-type or double mutant zebrafish kidney at 60 dpf (4 zebrafish kidneys and 2 independent repeats for each sample). Total RNA was extracted using TRIzol (Invitrogen). The concentrations and purity of the RNA samples were analyzed on a NanoDrop spectrophotometer (Isogen Life Science). The RNA integrity was confirmed on an Agilent 2100 Bioanalyzer (Agilent Technologies). Microarray was performed with Zebrafish Gene 1.0 ST Affymetrix Exon Array following the manufacturer’s instructions in Shanghai Biotechnology Co. Ltd. The ArrayExpress database accession number is E-MTAB-2692.
qPCR analysis
For the quantitative real-time polymerase chain reaction (qPCR) analysis of miRNA, specific primers were designed as described previously.33 The primers used for qPCR are listed in supplemental Table 3.
Results
Two miR-142 paralogs in zebrafish
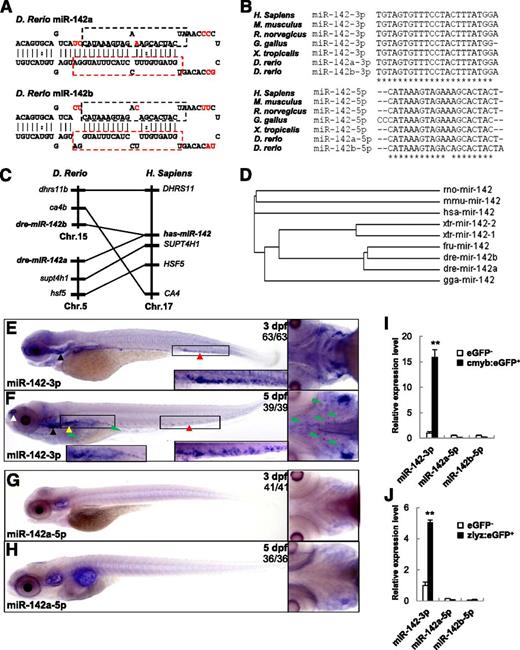
Unlike mammals, there are 2 paralogs of miR-142 in the zebrafish genome. One is located on chromosome 5, named miR-142a, and the other is located on chromosome 15, named miR-142b. Mfold analysis showed that miR-142a and miR-142b can fold into similar hairpin structures (Figure 1A), indicating that both can be processed into mature miRNAs from the 5′ and 3′ ends. miR-142a-3p and miR-142b-3p are identical in sequence (they were described as miR-142-3p in this paper); whereas miR-142a-5p differs from miR-142b-5p by only 1 nt, and the sequences are highly conserved in vertebrates (Figure 1B). Syntenic analysis showed that genes flanking zebrafish miR-142a/b were syntenic with the miR-142 locus in humans (Figure 1C). Combined with the phylogenic analysis (Figure 1D), these results suggest that a duplication event happened at the miR-142 loci in zebrafish.
Zebrafish miR-142-3p is enriched in myeloid cells and the definitive hematopoietic organ. (A) The stem-loop structure of pre-miR-142a and pre-miR-142b in zebrafish. The mature miR-142-3p sequence is boxed with a red dashed line. Mature miR-142a-5p and miR-142b-5p are boxed with a black dashed line. Different nucleotides are marked in red. (B) Multiple sequence alignment of miR-142-3p (top) and miR-142-5p (bottom) from different species. Fully conserved nucleotides are indicated with an asterisk. (C) Synteny of the miR-142 loci between the zebrafish and human. (D) Phylogenic analysis of miR-142. A phylogenic tree was constructed from a clustalW2 alignment using the neighbor-joining algorithm. (E-F) WISH with DIG-labeled LNA for miR-142-3p in zebrafish embryos at 3 dpf or 5 dpf. (A) miR-142-3p is specifically expressed in CHT (red triangle) and the thymus (black triangle) at 3 dpf. (B) miR-142-3p is expressed in CHT (red triangle), the kidney (yellow triangle), the thymus (black triangle), and the olfactory bulbs (white triangle) at 5 dpf. Expression of miR-142-3p in the tissue mesenchymes is indicated with a green triangle. (G-H) WISH for miR-142a-5p in zebrafish embryos at 3 dpf or 5 dpf. (I-J) qPCR analysis for mi-142-3p and miR-142a/b-5p in GFP-positive cells from Tg(cmyb:eGFP) (I) or Tg(zlyz:eGFP) (J) zebrafish at 3 dpf. **P < .01 vs GFP-negative cells; 2-tailed Student t test.
Zebrafish miR-142-3p is enriched in myeloid cells and the definitive hematopoietic organ. (A) The stem-loop structure of pre-miR-142a and pre-miR-142b in zebrafish. The mature miR-142-3p sequence is boxed with a red dashed line. Mature miR-142a-5p and miR-142b-5p are boxed with a black dashed line. Different nucleotides are marked in red. (B) Multiple sequence alignment of miR-142-3p (top) and miR-142-5p (bottom) from different species. Fully conserved nucleotides are indicated with an asterisk. (C) Synteny of the miR-142 loci between the zebrafish and human. (D) Phylogenic analysis of miR-142. A phylogenic tree was constructed from a clustalW2 alignment using the neighbor-joining algorithm. (E-F) WISH with DIG-labeled LNA for miR-142-3p in zebrafish embryos at 3 dpf or 5 dpf. (A) miR-142-3p is specifically expressed in CHT (red triangle) and the thymus (black triangle) at 3 dpf. (B) miR-142-3p is expressed in CHT (red triangle), the kidney (yellow triangle), the thymus (black triangle), and the olfactory bulbs (white triangle) at 5 dpf. Expression of miR-142-3p in the tissue mesenchymes is indicated with a green triangle. (G-H) WISH for miR-142a-5p in zebrafish embryos at 3 dpf or 5 dpf. (I-J) qPCR analysis for mi-142-3p and miR-142a/b-5p in GFP-positive cells from Tg(cmyb:eGFP) (I) or Tg(zlyz:eGFP) (J) zebrafish at 3 dpf. **P < .01 vs GFP-negative cells; 2-tailed Student t test.
Hematopoietic specific expression of miR-142-3p in zebrafish
To examine the spatial and temporal expression of miR-142-3p and miR-142a-5p, WISH was performed using LNA probes. miR-142-3p was widely expressed in zebrafish embryos within 24 hpf (supplemental Figure 1A-I). However, its expression was specifically restricted to caudal hematopoietic tissue (CHT) and the thymus at 3 dpf (Figure 1E). Finally at 5 dpf, miR-142-3p was highly enriched in all zebrafish definitive hematopoietic organs, including CHT, the kidney, and thymus, although expression signals could also be detected in some nasal sensory epithelium cells (Figure 1F). Notably, obvious expression of miR-142-3p could be detected in cells within the head and trunk tissue mesenchymes, which is similar to tissue-resident or interstitial macrophages.34 These results indicated that, as for mammals, miR-142-3p is a hematopoietic-specific miRNA in vertebrates.10,35
Interestingly, we could not detect any expression of miR-142a-5p by LNA-mediated WISH from the 1-cell stage to 5 dpf in zebrafish embryos (Figure 1G-H, some data not shown). Furthermore, qPCR assays were performed to validate the specific hematopoietic cell expression of miR-142-3p. The relative expression of miR-142-3p was 15-fold enriched in cmyb:eGFP-positive (cmyb:eGFP+) hematopoietic stem/progenitor cells (HSPCs), whereas the miR-142a/b-5p was almost undetectable (Figure 1I). Similarly, the relative expression of miR-142-3p was fivefold enriched in zlyz:eGFP-positive (zlyz:eGFP+) myelomonocytes, whereas miR-142a/b-5p was almost undetectable (Figure 1J). These results suggest that miR-142-3p, but not miR-142a/b-5p, plays an important role in zebrafish hematopoietic development.
ZFN-mediated targeted mutagenesis in miR-142a/b
To explore the function of miR-142-3p in vivo, we sought to inactivate miR-142a and miR-142b. Two candidate sites in miR-142a, which are potentially amenable to targeted mutagenesis using the OPEN ZFN method,26 were identified using the online ZiFiT software (supplemental Figure 2A-B). The spacers of the 2 targets were both near the seed region of miR-142a-3p. We carried out OPEN to engineer and select zinc-finger arrays targeted to the 4 9-bp half sites of the 2 candidate sites (supplemental Table 1). Finally, 1 selected pair of ZFNs could efficiently induce a variety of deletions or insertions within the target site (aCCCCTCGCCacagtGTAGTGTTTc) in zebrafish embryos (Figure 2A). The embryos injected with mRNA encoding the selected pair of ZFNs were raised and screened for founders with germ-line mutations in the miR-142a locus. After the outcross between the founders and wild-type zebrafish and then the intercross between the heterozygous F1 generation, we obtained a number homozygous miR-142a mutant lines for further investigation (supplemental Figure 3A).
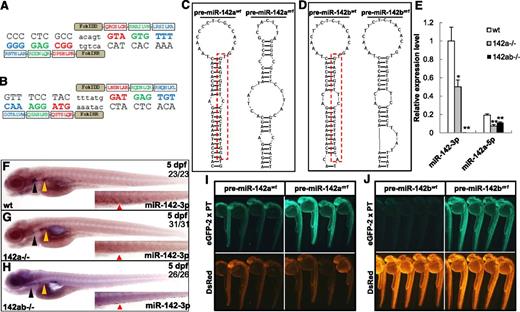
Deletion of miR-142-3p in miR-142a and miR-142b double mutant zebrafish. (A) Target sites in zebrafish miR-142a and amino acid sequences of the zinc-finger proteins were selected to recognize this site by OPEN. Spacer sequences in the target site are shown in lowercase letters. Each of the selected zinc-finger arrays targeting 9-bp nucleotides was fused with a subunit of the FokI nuclease. (B) The target site in zebrafish miR-142b and the amino acid sequences of the zinc-finger proteins were designed to recognize this site by CoDA. (C) Secondary structure prediction of the pre-miR-142a in the wild-type 142awt (left) or 142 am1 (right) allele. Mature wild-type miR-142-3p from pre-miR-142a is boxed with a red dashed line. (D) Secondary structure prediction of pre-miR-142b in wild-type miR-142bwt (left) or miR-142bm1 (right) allele. Mature wild-type miR-142-3p from pre-miR-142b is boxed with a red dashed line. (E) The expression level of miR-142-3p in wild-type, 142 am1/m1 (142a−/−), and 142 am1/m1142bm1/m1 (142ab−/−) mutant zebrafish embryos at 3 dpf, which was detected by real-time PCR. The expression of miR-142-3p and miR-142a-5p is normalized to glyceraldehyde-3-phosphate dehydrogenase (n = 3 for each genotype). Error bars indicate the standard error of the mean (SEM). **P < .01 compared with wild-type control. (F-H) Detection of miR-142-3p expression in wild-type, 142a−/−, and 142ab−/− mutant zebrafish embryos at 5 dpf by WISH. The black triangle indicates the thymus; yellow triangle, kidney; and red triangle, CHT. Note that the staining was weakened, but not diminished, in the 142a−/− zebrafish embryos. (I-J) In vivo reporter assays for the interaction between miR-142-3p and the targets with wild-type pre-miR-142a/bwt or mutant pre-miR-142a/bm1. EGFP mRNA harboring 2 perfect complementary target sites (EGFP-2 x PT) was coinjected with pre-miR-142awt (I), pre-miR-142 am1 (I), pre-miR-142bwt (J), or pre-miR-142bm1(J); DsRed mRNA served as a control.
Deletion of miR-142-3p in miR-142a and miR-142b double mutant zebrafish. (A) Target sites in zebrafish miR-142a and amino acid sequences of the zinc-finger proteins were selected to recognize this site by OPEN. Spacer sequences in the target site are shown in lowercase letters. Each of the selected zinc-finger arrays targeting 9-bp nucleotides was fused with a subunit of the FokI nuclease. (B) The target site in zebrafish miR-142b and the amino acid sequences of the zinc-finger proteins were designed to recognize this site by CoDA. (C) Secondary structure prediction of the pre-miR-142a in the wild-type 142awt (left) or 142 am1 (right) allele. Mature wild-type miR-142-3p from pre-miR-142a is boxed with a red dashed line. (D) Secondary structure prediction of pre-miR-142b in wild-type miR-142bwt (left) or miR-142bm1 (right) allele. Mature wild-type miR-142-3p from pre-miR-142b is boxed with a red dashed line. (E) The expression level of miR-142-3p in wild-type, 142 am1/m1 (142a−/−), and 142 am1/m1142bm1/m1 (142ab−/−) mutant zebrafish embryos at 3 dpf, which was detected by real-time PCR. The expression of miR-142-3p and miR-142a-5p is normalized to glyceraldehyde-3-phosphate dehydrogenase (n = 3 for each genotype). Error bars indicate the standard error of the mean (SEM). **P < .01 compared with wild-type control. (F-H) Detection of miR-142-3p expression in wild-type, 142a−/−, and 142ab−/− mutant zebrafish embryos at 5 dpf by WISH. The black triangle indicates the thymus; yellow triangle, kidney; and red triangle, CHT. Note that the staining was weakened, but not diminished, in the 142a−/− zebrafish embryos. (I-J) In vivo reporter assays for the interaction between miR-142-3p and the targets with wild-type pre-miR-142a/bwt or mutant pre-miR-142a/bm1. EGFP mRNA harboring 2 perfect complementary target sites (EGFP-2 x PT) was coinjected with pre-miR-142awt (I), pre-miR-142 am1 (I), pre-miR-142bwt (J), or pre-miR-142bm1(J); DsRed mRNA served as a control.
Because candidate target sites for OPEN ZFNs in miR-142b could not be identified, we generated heritable mutation of miR-142b in zebrafish using ZFNs engineered by the CoDA approach.27 One potential CoDA ZFN target site was predicted near the 3′ end of miR-142b-3p by the ZiFiT software (supplemental Figure 2C). CoDA ZFNs targeting this site were engineered (Figure 2B). We raised the embryos injected with the mRNA encoding the ZFNs and screened out a germ-line–transmitting founder with a 4-bp insertion at the 3′ end of miR-142b-3p. We obtained the homozygous miR-142b mutant line as described previously (supplemental Figure 3B). None of the homozygous miR-142a or miR-142b mutant zebrafish showed apparent defects in development, viability, or life span.
Deletion of redundant miR-142-3p in miR-142 double mutant zebrafish
To obtain the miR-142a and miR-142b double mutant zebrafish, homozygous miR-142am1/m1 (142a−/−) zebrafish were first crossed with homozygous miR-142bm1/m1 (142b−/−) and then double-heterozygous mutant zebrafish were intercrossed. Homozygous miR-142am1/m1miR-142bm1/m1 (142ab−/−) zebrafish were born at normal Mendelian ratios; they were fertile and displayed no obvious abnormalities. The 142ab−/− zebrafish possess 10-bp additions (Δ11 and +21) in both miR-142a loci (supplemental Figure 3A) and 4-bp additions in both miR-142b loci (supplemental Figure 3B). To assess the influence of the mutations within miR-142a or miR-142b on the miRNA maturation, we predicted the secondary structure of pre-miR-142am1 or pre-miR-142bm1 using the RNAStructure software (version 4.6). The normal stem-loop structures as in the wild-type pre-miR-142a were completely destroyed by the mutation in pre-miR-142am1 (Figure 2C), indicating that the miRNA maturation from the pre-miR-142am1 would be abolished. Compared with the normal stem-loop structure of pre-miR-142b, the 4-bp additions in pre-miR-142bm1 caused an additional bulb in the stem (Figure 2D).
Quantification by real-time PCR showed that the expression of miR-142-3p in 142a−/− embryos decreased by approximately half at 3 dpf, and no expression of miR-142-3p was detected in 142ab−/− zebrafish embryos (Figure 2E). Interestingly, the expression of miR-142a-5p was unexpectedly not abolished in either 142a−/− or 142ab−/− embryos, probably because miR-142b-5p was still processed from pre-miR-142bm1 and the primer for miR-142a-5p in qPCR could not discriminate the 1-nt difference between miR-142a-5p and miR-142b-5p. LNA-mediated WISH confirmed the results of the qPCR (Figure 2F-H).
In the target binding test, EGFP-2 x PT mRNA coinjected with pre-miR-142am1 or pre-miR-142bm1 was obviously derepressed compared with coinjection with wild-type pre-miR-142a or pre-miR-142b (Figure 2I-J), which suggested that mutations in pre-miR-142am or pre-miR-142bm1 efficiently abolished the function of miR-142-3p and derepressed its targets in vivo.
Deletion of miR-142-3p caused reduction in the number of neutrophils in fetal myelopoiesis and the involved mechanisms
We systematically investigated the definitive hematopoietic lineage in miR-142-3p–deficient embryos. WISH and qPCR showed normal expression of flk1 at 3 dpf, cmyb and gata1 at 3 dpf and 4 dpf, and rag1 at 5 dpf in 142ab−/− embryos, which respectively indicated normal angiogenesis in the CHT, unaffected homing of HSPCs and definitive erythropoiesis, and normal lymphopoiesis after depletion of miR-142-3p (supplemental Figure 4A-L).
However, WISH showed that cells expressing mpo, a specific neutrophil marker, were significantly reduced in number by ∼20% in the CHT of 142ab−/− embryos at 3 dpf (P < .01; Figure 3A-C). Consistently, the number of Sudan black positive (SB+) neutrophils reduced by 37% in 142ab−/− embryos (P < .01; Figure 3D-F). However, the SB+ neutrophils were normal in 142a−/− or 142b−/− embryos, indicating a redundant role of miR-142a-3p and miR-142b-3p in zebrafish neutrophil development (supplemental Figure 5A-E). Similarly, eGFP-labeled myelomonocytes in Tg(zlyz:eGFP) embryos were also affected by deletion of miR-142-3p. In 142ab−/−; Tg(zlyz:eGFP) embryos at 3 dpf, the zlyz:eGFP+ cells were reduced in number by 20% (P < .01; Figure 3G-I). Interestingly, we also observed that the mpo+ cells, SB+ cells, and zlyz:eGFP+ cells were abnormally scattered around the head in 142ab−/− embryos (Figure 3A’-H’).
Deletion of miR-142-3p caused reduction of neutrophils in myelopoiesis. (A-B) WISH for mpo in wild-type (A) and 142ab−/− (B) embryos at 3 dpf. (C) Quantification of mpo+ cell in wild-type and 142ab−/− embryos at 3 dpf. (D-E) Sudan black staining of neutrophils in wild-type (D) and 142ab−/− (E) embryos at 3 dpf. (F) Quantification of SB+ cell in wild-type and 142ab−/− embryos at 3 dpf. (G-H) Representative stereo fluorescence images of Tg(zlyz:eGFP) (G) and 142ab−/−; Tg(zlyz:eGFP) (H) embryos at 3 dpf. (I-J) Quantification of zlyz:eGFP+ cells in Tg(zlyz:eGFP) and 142ab−/−; Tg(zlyz:eGFP) embryos at 3 dpf. (I) zlyz:eGFP+ cells in the whole embryos; (J) zlyz:eGFP+ cells in the head or CHT. n ≥ 10; error bars represent SEM. (K) Quantification of zlyz:eGFP+ cells in Tg(zlyz:eGFP) and 142ab−/−; Tg(zlyz:eGFP) embryos of different stages, from 30 hpf to 96 hpf. n ≥ 10; error bars represent SEM. (L) Immunostaining shows the eGFP+ cells in the CHT of Tg(zlyz:eGFP) (upper panel) and 142ab−/−; Tg(zlyz:eGFP) (lower panel) embryos at 3 dpf. Macrophage-like cell is indicated by yellow triangle (lower panel). (M) WISH shows the mpeg1 expression in the CHT of wild-type (upper panel) and 142ab−/− embryos (lower panel) at 3 dpf. (N-S) Double immunostaining of zlyz:eGFP and BrdU shows a reduced proliferation of zlyz:eGFP+ cells in the CHT of 142ab−/− embryos at 3 dpf. (T) Quantification of the percentage of BrdU+ cells in the 3 dpf zlyz:eGFP+ population. n ≥ 8; error bars represent SEM. **P < .01; 2-tailed Student t test.
Deletion of miR-142-3p caused reduction of neutrophils in myelopoiesis. (A-B) WISH for mpo in wild-type (A) and 142ab−/− (B) embryos at 3 dpf. (C) Quantification of mpo+ cell in wild-type and 142ab−/− embryos at 3 dpf. (D-E) Sudan black staining of neutrophils in wild-type (D) and 142ab−/− (E) embryos at 3 dpf. (F) Quantification of SB+ cell in wild-type and 142ab−/− embryos at 3 dpf. (G-H) Representative stereo fluorescence images of Tg(zlyz:eGFP) (G) and 142ab−/−; Tg(zlyz:eGFP) (H) embryos at 3 dpf. (I-J) Quantification of zlyz:eGFP+ cells in Tg(zlyz:eGFP) and 142ab−/−; Tg(zlyz:eGFP) embryos at 3 dpf. (I) zlyz:eGFP+ cells in the whole embryos; (J) zlyz:eGFP+ cells in the head or CHT. n ≥ 10; error bars represent SEM. (K) Quantification of zlyz:eGFP+ cells in Tg(zlyz:eGFP) and 142ab−/−; Tg(zlyz:eGFP) embryos of different stages, from 30 hpf to 96 hpf. n ≥ 10; error bars represent SEM. (L) Immunostaining shows the eGFP+ cells in the CHT of Tg(zlyz:eGFP) (upper panel) and 142ab−/−; Tg(zlyz:eGFP) (lower panel) embryos at 3 dpf. Macrophage-like cell is indicated by yellow triangle (lower panel). (M) WISH shows the mpeg1 expression in the CHT of wild-type (upper panel) and 142ab−/− embryos (lower panel) at 3 dpf. (N-S) Double immunostaining of zlyz:eGFP and BrdU shows a reduced proliferation of zlyz:eGFP+ cells in the CHT of 142ab−/− embryos at 3 dpf. (T) Quantification of the percentage of BrdU+ cells in the 3 dpf zlyz:eGFP+ population. n ≥ 8; error bars represent SEM. **P < .01; 2-tailed Student t test.
Quantification of zlyz:eGFP+ cells indicated that the number of neutrophils in the CHT was decreased, whereas primitive neutrophils in the head were slightly increased (Figure 3J). Unexpectedly, the total number of 142ab−/−zlyz:eGFP+ cells were normal before 48 hpf, but started to decrease at 72 hpf (Figure 3K), indicating that deletion of miR-142-3p predominantly impaired the production of neutrophils in the CHT.
In further analysis of 142ab−/−zlyz:eGFP+ cells in the CHT by immunostaining, surprisingly, we observed abnormally hypermature tissue-resident macrophage-like cells in the trunk (Figure 3L). mpeg1+ macrophages were obviously reduced in the CHT of 142ab−/− embryos at 3 dpf (Figure 3M), suggesting that the fate of the missing neutrophils was not switched toward the macrophage program.
An additional 2 possibilities could account for the reduced neutrophil count in the CHT of 142ab−/− embryos, including of enhanced cell apoptosis and reduced cell proliferation. The terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling assay revealed no significant increase in the percentage of TUNEL+/zlyz:eGFP+ cells in the CHT of 142ab−/−; Tg(zlyz:eGFP) embryos (supplemental Figure 7A-G). The BrdU cooperation assay showed that ∼13% of zlyz:eGFP+ cells in the CHT of Tg(zlyz:eGFP) embryos were BrdU+, whereas only ∼3% of zlyz:eGFP+ cells in the mutant CHT were BrdU+ at 3 dpf. (P < .01; Figure 3N-T). Therefore, we reasoned that the reduction in the neutrophils in the CHT of 142ab−/− embryos was caused by the decreased proliferation of neutrophils instead of the enhanced apoptosis.
Hypermaturation and a dysregulated inflammatory response of neutrophils in 142ab−/− embryos
To further evaluate the neutrophil maturation, Wright-Giemsa staining of the myeloid zlyz:eGFP+ cells isolated by flow cytometry was performed. Surprisingly, we observed abnormal hypermaturation of neutrophils with bilobed and multilobed nuclei in 142ab−/− embryos (Figure 4A-B). There were ∼33% of zlyz:eGFP+ cells with bilobed nuclei and ∼14% ones with multilobed nuclei in 142ab−/− embryos, whereas there were only ∼15% of zlyz:eGFP+ cells with bilobed nuclei and rare cells with multilobed nuclei in wild-type embryos. Accordingly, the percentage of immature zlyz:eGFP+ cells in 142ab−/− embryos was significantly reduced. Further morphologic analysis revealed that the hypermature neutrophils with bilobed or multilobed nuclei showed enlarged cell size and a decreased nucleocytoplasmic ratio (Figure 4C-E). Collectively, those results indicated that deletion of miR-142-3p cause the hypermaturation of neutrophils in zebrafish myelopoiesis.
Aberrant hypermaturation of neutrophils in myelopoiesis in miR-142-3p–deficient embryos. (A-B) The percentage of immature, bilobed, and multilobed neutrophils in zlyz:eGFP+ cells. (A) Wild-type embryos (immature neutrophils 84.3%, standard deviation [SD] = 0.76; bibilobed neutrophils 14.6%, SD = 1.80; and multilobed neutrophils 1.1%, SD = 1.08). (B) The 142ab−/− embryos (immature neutrophils 53.2%, SD = 3.17; bibilobed neutrophils 32.9%, SD = 3.43; and multilobed neutrophils 13.9%, SD = 2.22). At least 100 cells were counted per experiment; SD, from 3 independent experiments. (C-E) Morphologic analysis of neutrophils in wild-type and 142ab−/− embryos at 3 dpf. zlyz:eGFP+ cells were sorted out from wild-type or 142ab−/− embryos and were stained with Wright-Giemsa (C). Hypersegmented neutrophils were indicated with red triangle. Relative cell size (D) and nucleocytoplasmic ratio (E) of wild-type and 142ab−/− neutrophils were quantified by ImageJ software (version 1.47). *P < .05 or **P < .01 vs wild-type; 2-tailed Student t test. Bar represents 8 μm. (F) Sudan black staining of neutrophils recruited to neuromasts in wild-type (upper panel) and 142ab−/− embryos (lower panel) after incubation with copper sulfate for 1.5 h. (G) Quantification of SB+ neutrophils recruited to neuromasts. n ≥ 15; error bars represent SEM. **P < .01 vs wild-type; 2-tailed Student t test.
Aberrant hypermaturation of neutrophils in myelopoiesis in miR-142-3p–deficient embryos. (A-B) The percentage of immature, bilobed, and multilobed neutrophils in zlyz:eGFP+ cells. (A) Wild-type embryos (immature neutrophils 84.3%, standard deviation [SD] = 0.76; bibilobed neutrophils 14.6%, SD = 1.80; and multilobed neutrophils 1.1%, SD = 1.08). (B) The 142ab−/− embryos (immature neutrophils 53.2%, SD = 3.17; bibilobed neutrophils 32.9%, SD = 3.43; and multilobed neutrophils 13.9%, SD = 2.22). At least 100 cells were counted per experiment; SD, from 3 independent experiments. (C-E) Morphologic analysis of neutrophils in wild-type and 142ab−/− embryos at 3 dpf. zlyz:eGFP+ cells were sorted out from wild-type or 142ab−/− embryos and were stained with Wright-Giemsa (C). Hypersegmented neutrophils were indicated with red triangle. Relative cell size (D) and nucleocytoplasmic ratio (E) of wild-type and 142ab−/− neutrophils were quantified by ImageJ software (version 1.47). *P < .05 or **P < .01 vs wild-type; 2-tailed Student t test. Bar represents 8 μm. (F) Sudan black staining of neutrophils recruited to neuromasts in wild-type (upper panel) and 142ab−/− embryos (lower panel) after incubation with copper sulfate for 1.5 h. (G) Quantification of SB+ neutrophils recruited to neuromasts. n ≥ 15; error bars represent SEM. **P < .01 vs wild-type; 2-tailed Student t test.
In zebrafish, neutrophils were quickly recruited to stressed tissues in the acute inflammatory response.36-38 We thus investigated the inflammatory response of miR-142-3p–deficient neutrophils in vivo. In a copper sulfate–induced inflammatory assay, infiltration of neutrophil to neuromasts was significantly weakened in miR-142-3p–deficient embryos (Figure 4F-G). Additionally, in 142ab−/− zebrafish embryos at 3 dpf, neutrophils recruited to the wound at 6 h post–tail transection significantly decreased compared with wild-type embryos (supplemental Figure 8A-C). These data indicated an impaired chemotactic migration of the neutrophil response to inflammation by the deletion of miR-142-3p. Central to the function of neutrophils is their ability to recognize and phagocytose microbes. Analysis of phagocytosis showed the normal ability for bacteria ingestion by 142ab−/−zlyz:eGFP+ cells (supplemental Figure 8D-F). Collectively, these results suggested an important role for miR-142-3p in regulating the inflammatory migration of neutrophils, but miR-142-3p did not seem to be involved in neutrophil phagocytosis.
Deletion of miR-142-3p caused hypermaturation of neutrophils in zebrafish adult myelopoiesis
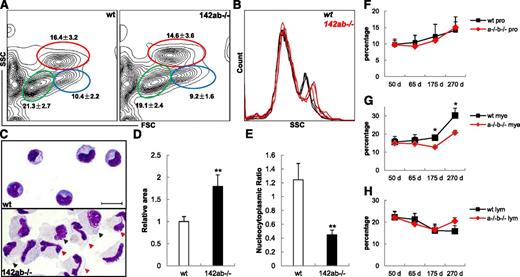
We also analyzed the adult hematopoietic hierarchy in miR-142-3p–deficient kidneys, the main adult hematopoietic site in zebrafish. The percentage of each hematopoietic cell lineage fraction in 142ab−/− WKM is similar to that in wild type, indicating that the normal lineage-specific differentiation of the long-term hematopoietic stem cell (HSCs) was unaffected by the deletion of miR-142-3p (Figure 5A). However, the granularity of the myelomonocytes by the side scatter analysis was increased obviously in the 142ab−/− WKM (Figure 5B). Morphology analysis of the zlyz:eGFP+ myelomonocytes39 in 142ab−/− kidneys showed aberrant hypermaturation of neutrophils, irregular cell shape, increased and enlarged cell size, decreased nucleocytoplasmic ratio, and unusual mature macrophage-like cells (Figure 5C-E). In contrast to the aberrant hypermaturation of neutrophils in 142ab−/− kidneys, eosinophils in the zlyz:eGFP- myelomonocytic fraction were almost unaffected as indicated by morphologic analysis (supplemental Figure 9A). In addition, no obvious difference was observed in erythrocytes between the 142ab−/− and wild-type kidneys (supplemental Figure 9B).
Aberrant hypermaturation of neutrophils in zebrafish adult kidney hematopoiesis after deletion of miR-142-3p. (A) Flow cytometry analysis of WKM in wild-type (left panel) and 142ab−/− (right panel) zebrafish. Myelomonocytes (mye) were indicated in red circle; hematopoietic progenitors (pro), blue circle; and lymphocytes and lymphocyte-like cells (lym), green circle. Average percentage ± SD for each fraction from 8 individual zebrafish at 65 dpf was shown. (B) Comparison of side scatter analysis (SSC) of wild-type and 142ab−/− WKMs. Data were from 3 individual zebrafish in each group. (C) Wright-Giemsa staining of lyz-GFP+ myelomonocytes in wild-type (upper panel) or 142ab−/− (lower panel) zebrafish kidney. Hypermature neutrophils were indicated with red triangle. Macrophage-like cells were indicated with black triangle. Bar represents 8 μm. (D-E) Quantification of cell size (D) and nucleocytoplasmic ratio (E) of neutrophils from wild-type and 142ab−/− zebrafish kidney. (F-H) Quantification of the 3 hematopoietic fractions in wild-type and 142ab−/− kidney with age. Hematopoietic progenitors (F), myelomonocytes (G), and lymphocytes and lymphocyte-like cells (H). WKM from at least 4 individuals was analyzed by flow cytometry in each group.*P < .05 or **P < .01 vs wild-type; 2-tailed Student t test.
Aberrant hypermaturation of neutrophils in zebrafish adult kidney hematopoiesis after deletion of miR-142-3p. (A) Flow cytometry analysis of WKM in wild-type (left panel) and 142ab−/− (right panel) zebrafish. Myelomonocytes (mye) were indicated in red circle; hematopoietic progenitors (pro), blue circle; and lymphocytes and lymphocyte-like cells (lym), green circle. Average percentage ± SD for each fraction from 8 individual zebrafish at 65 dpf was shown. (B) Comparison of side scatter analysis (SSC) of wild-type and 142ab−/− WKMs. Data were from 3 individual zebrafish in each group. (C) Wright-Giemsa staining of lyz-GFP+ myelomonocytes in wild-type (upper panel) or 142ab−/− (lower panel) zebrafish kidney. Hypermature neutrophils were indicated with red triangle. Macrophage-like cells were indicated with black triangle. Bar represents 8 μm. (D-E) Quantification of cell size (D) and nucleocytoplasmic ratio (E) of neutrophils from wild-type and 142ab−/− zebrafish kidney. (F-H) Quantification of the 3 hematopoietic fractions in wild-type and 142ab−/− kidney with age. Hematopoietic progenitors (F), myelomonocytes (G), and lymphocytes and lymphocyte-like cells (H). WKM from at least 4 individuals was analyzed by flow cytometry in each group.*P < .05 or **P < .01 vs wild-type; 2-tailed Student t test.
Although the homozygous 142ab−/− zebrafish could be grown up without apparent abnormalities, only 20% of the 142ab−/−zebrafish survived to 2 years of age (data not shown). Flow cytometry-based lineage analysis showed that the percentage of myelomonocytic fraction, but not the progenitor or lymphocytes in 142ab−/− adult kidneys, decreased gradually with age (Figure 5F-H), which might suggest a chronic failure in adult myelopoiesis that is caused by deletion of miR-142-3p.
Dysregulated transcriptome in zebrafish myelopoiesis by deletion of miR-142-3p
To further investigate how the deletion of miR-142-3p caused aberrant hypermaturation in neutrophil development in both the fetal stage and adult kidney, we performed an Affymetrix microarray analysis of the adult zebrafish kidney. Transcriptome analysis revealed 2458 differentially expressed probe sets (>1.5-fold change) in 142ab−/− myelomonocytes (Figure 6A). Combined with transcriptome analysis of WKM between 142ab−/− and wild-type zebrafish, 518 transcripts were identified as obviously coupregulated, whereas 529 transcripts were obviously codownregulated (Figure 6A), both in myelomonocytes and WKM, by the deletion of miR-142-3p. Bioinformatic prediction of targets for miR-142-3p was performed within the 1047 codysregulated transcripts (supplemental Figure 12). Putative miR-142-3p target sites were significantly enriched in the fraction of transcripts that were coupregulated compared with the codownregulated fraction (Figure 6B).
Deletion of miR-142-3p caused trancriptome changes and activated interferon (IFN) γ signaling in zebrafish kidney myelopoiesis. (A) The difference and overlap between differential expressed genes (1.5-fold) in WKM after deletion of miR-142-3p and that in myelomonocytes. (B) The distribution of number of candidate target sites for miR-142-3p in codysregulated genes (twofold change) in WKM (without erythrocytes) and myelomonocytes after deletion of miR-142-3p. (C) Comparison of codysregulated IFN-γ signaling–related genes both in 142ab−/− WKM (without erythrocytes) and 142ab−/− myelomonocytes (1.5-fold). (D) qPCR analysis of expression of IFN-γ signaling–related genes in wild-type and 142ab−/− myelomonocytes in zebrafish kidney. *P < .05 or **P < .01 vs wild-type; 2-tailed Student t test.
Deletion of miR-142-3p caused trancriptome changes and activated interferon (IFN) γ signaling in zebrafish kidney myelopoiesis. (A) The difference and overlap between differential expressed genes (1.5-fold) in WKM after deletion of miR-142-3p and that in myelomonocytes. (B) The distribution of number of candidate target sites for miR-142-3p in codysregulated genes (twofold change) in WKM (without erythrocytes) and myelomonocytes after deletion of miR-142-3p. (C) Comparison of codysregulated IFN-γ signaling–related genes both in 142ab−/− WKM (without erythrocytes) and 142ab−/− myelomonocytes (1.5-fold). (D) qPCR analysis of expression of IFN-γ signaling–related genes in wild-type and 142ab−/− myelomonocytes in zebrafish kidney. *P < .05 or **P < .01 vs wild-type; 2-tailed Student t test.
Sixty-four putative target sites with consecutive 8-nt sequences complementary to the 5′ seed region of miR-142-3p were identified in the full transcripts of 56 genes (supplemental Table 2). Four putative target sites for miR-142-3p in the 3′ untranslated region of waslb (2 sites) and abhd14b (2 sites) were confirmed to be efficiently inhibited by miR-142-3p in vivo (supplemental Figure 10A-F). However, the decreased number of neutrophils in the miR-142-3p–deficient zebrafish could not be rescued by morpholino-mediated knockdown of waslb or/and abhd14b (supplemental Figure 11A-F).
Activation of IFN-γ signaling mediated the impaired neutrophil development in the case of miR-142-3p deletion
We then analyzed the 518 coupregulated transcripts using the KEGG Pathway Database and found that the Janus kinase–signal transducer and activator of transcription (STAT) pathway was listed at the top of the significantly enriched pathways. As an extensive signaling that is downstream of cytokine receptors, the Janus kinase–STAT signaling pathway could be activated by a number of different ligands, including IFN-α/β, IFN-γ, epidermal growth factor, interleukin 6, and granulocyte colony-stimulating factor.40-42 Detailed analysis revealed that a series of IFN-γ–regulated genes were aberrantly upregulated in 142ab−/− myelomonocytes, consistent with qPCR results (Figure 6C). For instance, crfb17, a necessary component of IFN-γ receptor complexes in zebrafish, was upregulated by 6.35-fold.43 We also identified obvious upregulation of 2 interferon regulatory factor (IRF) family members, irf1b (2.92-fold) and irf9 (2.15-fold) (Figure 6D).
To test whether the activation of IFN-γ signaling is involved in impaired neutrophil development by deletion of miR-142-3p, morpholinos targeting stat1a, stat1b, or irf1b were injected into 142ab−/− embryos. We observed that knockdown of stat1a could partially rescue the decrease in neutrophils, whereas knockdown of stat1b or irf1b showed no rescue effect. However, surprisingly, combined knockdown of stat1a and irf1b in 142ab−/− embryos could completely reverse the decrease in neutrophils (Figure 7A-G). By further morphologic analysis, this combined knockdown of stat1a and irf1b dramatically reduced the percentage of hypersegmented (2.5%) or bilobed (23.8%) neutrophils in zlyz:eGFP+ cells in 142ab−/− embryos to the level of wild-type embryos (Figure 7H-P). And, there was also a relatively significant reduction in the percentage of hypermature neutrophils in 142ab−/−zlyz:eGFP+ cells by knockdown of stat1a (hypersegmented neutrophils, 7.6%; bilobed neutrophils, 29.5%). Interestingly, the impaired inflammatory migration of 142ab−/− neutrophils could also be partially rescued by the combined knockdown of stat1a and irf1b (Figure 7Q). Collectively, these results indicate that aberrant activation of the IFN-γ signaling, especially the upregulation of stat1a and irf1b, mediated the abnormality of neutrophils by deletion of miR-142-3p.
Abnormal activation of IFN-γ signaling attributed to the impaired development of neutrophils in 142ab−/− mutants. (A-F) Sudan black staining of neutrophils in wild-type embryos (A) and 142ab−/− embryos (B) and 142ab−/− embryos injected with stat1a MO (C), irf1b MO (D), stat1a MO and irf1b MO (E), and stat1b MO (F). (G) Quantification of SB+ cells in whole embryos. *P < .05 or **P < .01; 2-tailed Student t test. (H-K) Morphologic analysis of zlyz:eGFP+ cells from 3-dpf embryos. zlyz:eGFP+ cells were sorted from Tg(zlyz:eGFP) embryos (H); 142ab−/−; Tg(zlyz:eGFP) embryos (I); 142ab−/−; Tg(zlyz:eGFP) embryos injected with stat1a MO (J); or 142ab−/−; Tg(zlyz:eGFP) embryos coinjected with stat1a MO and irf1b MO (K) at 3 dpf and stained with Wright-Giemsa solution. (L-O) The percentage of immature, bilobed, and multilobed neutrophils in zlyz:eGFP+ cells. (L) Tg(zlyz:eGFP) embryos (immature neutrophils 81.2%, SD = 2.20; bibilobed neutrophils 17.2%, SD = 2.50; and multilobed neutrophils 1.6%, SD = 1.14). (M) The 142ab−/− embryos (immature neutrophils 55.7%, SD = 2.43; bibilobed neutrophils 31.1%, SD = 2.26; and multilobed neutrophils 13.2%, SD = 1.78). (N) The 142ab−/−; Tg(zlyz:eGFP) embryos injected with stat1a MO (immature neutrophils 62.9%, SD = 3.82; bibilobed neutrophils 29.5%, SD = 3.17; and multilobed neutrophils 7.6%, SD = 1.01). (O) The 142ab−/− ; Tg(zlyz:eGFP) embryos coinjected with stat1a MO and irf1b MO (immature neutrophils 73.8%, SD = 2.32; bibilobed neutrophils 23.8%, SD = 2.56; and multilobed neutrophils 2.5%, SD = 0.47). At least 100 cells were counted per experiment; SD, from 3 independent experiments. (P) Comparison of the percentage of bilobed or multilobed neutrophils in zlyz:eGFP+ cells. *P < .05 or **P < .01 vs wild-type; 2-tailed Student t test. (Q) Quantification of SB+ neutrophils recruited to neuromasts. *P < .05 or **P < .01 vs wild-type; 2-tailed Student t test.
Abnormal activation of IFN-γ signaling attributed to the impaired development of neutrophils in 142ab−/− mutants. (A-F) Sudan black staining of neutrophils in wild-type embryos (A) and 142ab−/− embryos (B) and 142ab−/− embryos injected with stat1a MO (C), irf1b MO (D), stat1a MO and irf1b MO (E), and stat1b MO (F). (G) Quantification of SB+ cells in whole embryos. *P < .05 or **P < .01; 2-tailed Student t test. (H-K) Morphologic analysis of zlyz:eGFP+ cells from 3-dpf embryos. zlyz:eGFP+ cells were sorted from Tg(zlyz:eGFP) embryos (H); 142ab−/−; Tg(zlyz:eGFP) embryos (I); 142ab−/−; Tg(zlyz:eGFP) embryos injected with stat1a MO (J); or 142ab−/−; Tg(zlyz:eGFP) embryos coinjected with stat1a MO and irf1b MO (K) at 3 dpf and stained with Wright-Giemsa solution. (L-O) The percentage of immature, bilobed, and multilobed neutrophils in zlyz:eGFP+ cells. (L) Tg(zlyz:eGFP) embryos (immature neutrophils 81.2%, SD = 2.20; bibilobed neutrophils 17.2%, SD = 2.50; and multilobed neutrophils 1.6%, SD = 1.14). (M) The 142ab−/− embryos (immature neutrophils 55.7%, SD = 2.43; bibilobed neutrophils 31.1%, SD = 2.26; and multilobed neutrophils 13.2%, SD = 1.78). (N) The 142ab−/−; Tg(zlyz:eGFP) embryos injected with stat1a MO (immature neutrophils 62.9%, SD = 3.82; bibilobed neutrophils 29.5%, SD = 3.17; and multilobed neutrophils 7.6%, SD = 1.01). (O) The 142ab−/− ; Tg(zlyz:eGFP) embryos coinjected with stat1a MO and irf1b MO (immature neutrophils 73.8%, SD = 2.32; bibilobed neutrophils 23.8%, SD = 2.56; and multilobed neutrophils 2.5%, SD = 0.47). At least 100 cells were counted per experiment; SD, from 3 independent experiments. (P) Comparison of the percentage of bilobed or multilobed neutrophils in zlyz:eGFP+ cells. *P < .05 or **P < .01 vs wild-type; 2-tailed Student t test. (Q) Quantification of SB+ neutrophils recruited to neuromasts. *P < .05 or **P < .01 vs wild-type; 2-tailed Student t test.
Discussion
Emerging evidence shows that miRNAs are substantially involved in myelopoiesis, but the role of miRNAs in the development of neutrophils remains elusive. In this study, we provided genetic evidence that miR-142-3p plays an essential role in the regulation of neutrophil homeostasis and maturation. Up to this point, the best-known miRNA that regulates neutrophil development is miR-223. A previous study in knockout mouse has revealed that miR-223 acts as a negative regulator of progenitor proliferation and granulocyte differentiation.7 As revealed by that study, miR-223–deficient neutrophils exhibit an unusual hypermature morphology that is characterized by nuclear hypersegmentation and blebbing, which is reminiscent of the granulocytes observed in human myelokathexis,44 and those abnormalities of neutrophils were also observed in our miR-142ab−/− knockout zebrafish. Moreover, a previous study showed that miR-142 is positively regulated by miR-223 in the myeloid cell line K562. Considering the previous evidence and our data, we speculated that miR-142-3p may be a key regulator in the common pathway of neutrophil maturation. Additionally, the 142ab−/− zebrafish could be a feasible animal model for the investigation of congenital neutropenia in human.
The function of miR-142-3p remains controversial because investigators focus on different developmental stages or the cell population in myelopoiesis. The expression of miR-142-3p was abolished in all of the neutrophil populations in our study, whereas miR-142-3p in the previous study was overexpressed in the all-trans-retinoic acid-treated NB4 leukemia cell line,13 which resembles a very early stage granulocyte progenitor. In addition, a previous study in knockout mouse reported that the genetic differentiation program for classic CD4+ dendritic cell development is not affected by deletion of miR-142-3p,15 which, combined with our results, suggests that overexpression strategies might cause a distinct consequence compared with the deletion studies.
To the best of our knowledge, functional studies of single miRNA in zebrafish were previously limited to morpholino- or LNA-mediated knockdown. Using ZFN-mediated targeted mutagenesis, we established the first miRNA double knockout zebrafish line and identified the redundant role of miR-142-3p processed from the 2 miR-142 paralogs in the regulation of normal neutrophil maturation and homeostasis. However, a recent knockdown study reported the obvious defects in HSC formation and T-cell differentiation,45 which was not observed in our 142ab−/− double mutant zebrafish. In our analysis of hematopoietic development in miR-142-3p morphants, we observed wide defects in hematopoiesis, including the cmyb+ HSPCs in CHT and the flk1+ vascular integrity. Notably, the defects in HSPC development and vascular integrity could be efficiently rescued by combined knockdown of p53 and miR-142-3p (supplemental Figure 6A-F), suggesting that a wide activation of p53 might also contribute to these defects. Considering the possibility of sequence-specific p53 activation induced by morpholino,46 combined knockdown of p53 would be helpful for the careful analysis of the HSC defects in miR-142-3p morphants.
Fetal definitive hematopoiesis in zebrafish begins in the ventral wall of dorsal aorta and then shifts to CHT, an intermediate hematopoietic site responsible for the proliferation and differentiation of HSPCs. Finally, long-term definitive hematopoiesis is established in the kidney along with HSPCs that colonize there at 5 dpf.16 In this study, deletion of miR-142-3p in zebrafish led to aberrant hypermaturation of neutrophils in both embryos and adult kidneys, which emphasized a consistent regulatory role of miR-142-3p in both fetal and adult hematopoiesis. However, the significant decrease of neutrophils in the fetal stage attributable to deletion of miR-142-3p could not be observed in adult kidneys before 3 months following fertilization. This may suggest a difference in the proliferation of neutrophil precursors after the deletion of miR-142-3p, which is in accord with the previous description that there may be a distinction in definitive hematopoiesis between the fetal stage and adult kidneys.47
Whole-transcriptome analysis of the myelomonocytic fraction in miR-142-3p–deficient kidneys revealed widely abnormal activation of IFN-γ signaling. We provided evidence that upregulation of stat1a and irf1b was mainly responsible for the impaired neutrophils. However, knockdown of irf1b could not reverse the decreased number of neutrophils in 142ab−/− embryos. Considering the antiproliferation effect of STAT1α, the upregulation of irf1b might be mainly responsible for the abnormal maturation of neutrophils. Notably, no potential binding sites were identified in the 3′ untranslated region of stat1a and irf1b, which suggested they were indirectly regulated by miR-142-3p. Interestingly, several studies in models of human autoimmune disorders implicated the suppressive effects of IFN-γ signaling on inflammatory neutrophil infiltration,48-51 which might partially explain the impaired response to acute inflammation of miR-142-3p–deficient neutrophils observed in this study.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Len Zon at Harvard Medical School for providing the Tg(cmyb:eGFP) line; Dr Yi Zhou at Children’s Hospital in Boston, Dr Thomas Look at Harvard Medical School, and Dr Keith Joung at Massachusetts General Hospital-East for helpful discussions and providing important suggestions; and Jiang Zhu, Wu Zhang, Xiang Miao, and Wen-wen Liu for technical assistance in flow cytometry. The authors are also grateful to all the members in the Laboratory of Development and Diseases for helpful discussions.
This work was supported by grants from the National Basic Research Program of China (2007CB947003), the Strategic Priority Research Program of the Chinese Academy of Science (XDA 01010109), the National Natural Science Foundation of China (81172694, 31322038, and 81370232), the Research Project of Chinese Ministry of Education (213015A), the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD) and Qinglan project (JX2161015124), the Outstanding Youth Fund of Jiangsu Province (SBK2014010296), and the Pujiang foundation of Shanghai (12PJ1410200).
Authorship
Contribution: H.-B.F. and Y.-J.L. performed experiments and analyzed data; L.W., T.-T.D., M. Dong, Y.J., Y.C., L.G., Z.-Z.M, and M. Deng assisted with experiments; H.-B.F., M. Deng, Q.J., H.-T.Y., A.-H.G., T.-X.L., and Y.Z. designed the research plan; and H.-B.F., Q.J., A.-H.G., and Y.Z. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Ting-Xi Liu died on July 16, 2011.
Correspondence: Aihua Gu, State Key Laboratory of Reproductive Medicine, Institute of Toxicology, School of Public Health, Nanjing Medical University, Nanjing 210029, China; e-mail: aihuagu@njmu.edu.cn; and Yong Zhou, Institute of Health Sciences, Chinese Academy of Sciences, Room 408, Building 1, 225 South Chong Qing Rd, Shanghai 200025, China; e-mail: zhouyong@sibs.ac.cn.
References
Author notes
H.-B.F. and Y.-J.L. contributed equally to this study.


![Figure 4. Aberrant hypermaturation of neutrophils in myelopoiesis in miR-142-3p–deficient embryos. (A-B) The percentage of immature, bilobed, and multilobed neutrophils in zlyz:eGFP+ cells. (A) Wild-type embryos (immature neutrophils 84.3%, standard deviation [SD] = 0.76; bibilobed neutrophils 14.6%, SD = 1.80; and multilobed neutrophils 1.1%, SD = 1.08). (B) The 142ab−/− embryos (immature neutrophils 53.2%, SD = 3.17; bibilobed neutrophils 32.9%, SD = 3.43; and multilobed neutrophils 13.9%, SD = 2.22). At least 100 cells were counted per experiment; SD, from 3 independent experiments. (C-E) Morphologic analysis of neutrophils in wild-type and 142ab−/− embryos at 3 dpf. zlyz:eGFP+ cells were sorted out from wild-type or 142ab−/− embryos and were stained with Wright-Giemsa (C). Hypersegmented neutrophils were indicated with red triangle. Relative cell size (D) and nucleocytoplasmic ratio (E) of wild-type and 142ab−/− neutrophils were quantified by ImageJ software (version 1.47). *P < .05 or **P < .01 vs wild-type; 2-tailed Student t test. Bar represents 8 μm. (F) Sudan black staining of neutrophils recruited to neuromasts in wild-type (upper panel) and 142ab−/− embryos (lower panel) after incubation with copper sulfate for 1.5 h. (G) Quantification of SB+ neutrophils recruited to neuromasts. n ≥ 15; error bars represent SEM. **P < .01 vs wild-type; 2-tailed Student t test.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/124/8/10.1182_blood-2013-12-545012/4/m_1320f4.jpeg?Expires=1769092295&Signature=SvOuy14349tm6Iuae4FfU9R3ickIzgzQue3E9fHrz-x7yBdrWsfVUc~LLt3gyApCT0DCa-yT9~FsBkLHyJVVRxGQpt5q4TFIcQSAlZmk76t~BVXL9JtNyCWeS0i9gozUq75Xak~RcMPlLfl5ylYjROLGcLKbh0JoVv17CI1-VILDJH4tiSJxulWFB6EgbpkKwMnNrI-eX4rJHBUzukgeMwubMuS393H6oLGXbpmpS5yBo2KLOOkWZUSCE6Ni9l-tnyUkrsdqO1~JxKtZuhV5jlrpMGR1OqNIn~hqwPOA6emMiqbvkIRVSvgOW23n8GsmuOU-5ru2d-HcwfQbF5CP8g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal