Key Points
We identified CRBN-binding proteins and confirmed recent work implicating the Ikaros transcription factors as important members.
Low IZKF1 levels predict lack of IMiD responsiveness and shorter OS; IZKF1 expression is a potential predictive biomarker for clinic application.
Abstract
Cereblon (CRBN) mediates immunomodulatory drug (IMiD) action in multiple myeloma (MM). Using 2 different methodologies, we identified 244 CRBN binding proteins and established relevance to MM biology by changes in their abundance after exposure to lenalidomide. Proteins most reproducibly binding CRBN (>fourfold vs controls) included DDB1, CUL4A, IKZF1, KPNA2, LTF, PFKL, PRKAR2A, RANGAP1, and SHMT2. After lenalidomide treatment, the abundance of 46 CRBN binding proteins decreased. We focused attention on 2 of these—IKZF1 and IKZF3. IZKF expression is similar across all MM stages or subtypes; however, IKZF1 is substantially lower in 3 of 5 IMiD-resistant MM cell lines. The cell line (FR4) with the lowest IKZF1 levels also harbors a damaging mutation and a translocation that upregulates IRF4, an IKZF target. Clinical relevance of CRBN-binding proteins was demonstrated in 44 refractory MM patients treated with pomalidomide and dexamethasone therapy in whom low IKZF1 gene expression predicted lack of response (0/11 responses in the lowest expression quartile). CRBN, IKZF1, and KPNA2 levels also correlate with significant differences in overall survival. Our study identifies CRBN-binding proteins and demonstrates that in addition to CRBN, IKZF1, and KPNA2, expression can predict survival outcomes.
Introduction
Immunomodulatory drugs (IMiDs) are key components of treatment of a number of hematological malignancies.1 The first drug in this class, thalidomide, became infamous for its teratogenic effects. Early mechanism of action studies implicated multiple potential, but no definitive drug activities, that explained this teratogenicity.2,3 Recently, however, Ito et al described that cereblon (CRBN) is the primary target of thalidomide teratogenicity.4 Thalidomide binds CRBN to alter the function of the E3 ubiquitin ligase complex (composed of CRBN, DDB1, and CUL4). Drug-induced downstream effects of altered CRBN activity include cell-cycle arrest with upregulation of the cyclin-dependent kinase inhibitor p21WAF-15 and downregulation of interferon regulatory factor 4 (IRF4), a MM cell survival factor that targets critical genes including MYC, CDK6, and CASP.6-8 Collateral effects on immune function including inhibition of tumor necrosis factor and upregulation of interleukin 2, T lymphocytes, and natural killer cells are also documented.
We and others previously reported that CRBN is also required for the anti-MM action of the thalidomide derivatives lenalidomide and pomalidomide, more accurately referred to collectively as “cereblon-binding small molecules.”9 Furthermore, we observed that CRBN expression decreases in MM patients who developed resistance to lenalidomide therapy.9,10 Conversely, loss of CRBN expression did not affect response to other agents, such as bortezomib, dexamethasone, and melphalan.9 Recent clinical correlative studies have observed a positive association between CRBN and response to thalidomide maintenance and up-front lenalidomide and dexamethasone therapy.11,12 Furthermore, we have observed CRBN and IRF4 mutations in drug-refractory patients, supporting the key role of CRBN in the response to IMiDs.13,14 Finally, we have reported that CRBN expression is highly predictive of response and survival outcomes after pomalidomide therapy.15 However, a majority of patients with low CRBN levels have no evidence of genomic mutation, thus transcriptional or posttranscriptional factors may influence CRBN gene expression and responsiveness to IMiD therapy.16
Despite these findings, the intermediaries and facilitators of cell death after CRBN and IMiD interaction that ultimately degrade IRF4 and MYC are as yet not fully elucidated. In the current study, we set out to identify CRBN-binding proteins and then determine which of these proteins were relevant to MM cytotoxicity by assessing alterations in abundance of CRBN-binding partners in the presence of lenalidomide. In the interim, 2 recent papers described that the transcription factors Ikaros (IKZF1) and Aiolos (IKZF3) were important intermediaries that are rapidly ubiquitinated and degraded after IMiDs bind to CRBN.17,18 We describe our identification of these, but also other binding proteins of relevance, and the correlation of these proteins with response and survival outcomes in MM patients.
Methods
Cells and reagents
Human myeloma cell lines (HMCLs) were maintained in RPMI-1640 medium, supplemented with 5% of sterile fetal calf serum and antibiotics. Anti-CRBN antibodies were obtained from Sigma-Aldrich (St Louis, MO); anti-IRF4, anti-PARP, anti-KPNA2, and anti-DDB1 were obtained from Cell Signaling Technology (Danvers, MA); anti-IKZF1 antibodies were obtained from Cell Signaling Technology and Santa Cruz Biotechnology (Dallas, TX); anti-IKZF3 was obtained from Imgenex (San Diego, CA); anti-MYC antibody was obtained from Epitomics (Burlingame, CA); lenalidomide was obtained from LC Laboratories (Woburn, MA); and Ni-NTA agarose was obtained from Qiagen (Valencia, CA).
All experiments were conducted under approval of the institutional review board (IRB2207-02). Patient samples were collected and stored under IRB919-04. This study was conducted in accordance with the Declaration of Helsinki.
Preparation of lentiviral-expressing virus and infection of myeloma cells
Human CRBN, IKZF1, and IKZF3 cDNAs were purchased from Thermo Scientific (Rockford, IL) and were subcloned into a lentiviral expression vector, pCDH-CMV-MCS-EF1-copGFP (System Bioscience, Mountain View, CA). CRBN tagged with His at its C-terminal was generated by polymerase chain reaction and confirmed by sequencing. IKZF1 and IKZF3 lentiviral shRNA constructs were purchased from Sigma-Aldrich. Lentivirus harboring control vector and all expression constructs were used to infect HMCLs.19 The infection efficiency was measured by FACScan analysis of green fluorescent protein (GFP) expression at day 3 after infection. OCI-MY5 cells infected by control virus (vector alone) and CRBN-expressing virus were sorted for GFP expression at 14 days after infection. Overexpression of CRBN was confirmed by immunoblotting assay.
Identification of CRBN binding proteins using coimmunoprecipitation (Co-IP) and Ni Charged (Ni+) beads
OCI-MY5 cells stably expressing vector alone (OCI-MY5/vec) or His-tagged CRBN (OCI-MY5/CRBN-His) were harvested and processed using 2 different pull-down methods:
Co-IP with an anti-CRBN antibody. OCI-MY5/CRBN-His cells were lysed using lysis buffer (50 mM Tris pH 8.0, 150 mM NaCl, 1% NP40, 0.05% Tween20, and proteinase inhibitors) and protein quantified using a bicinchoninic acid assay. 1 mg of protein was precleaned by incubation with protein A/G slurry at 4°C with rocking, and resultant supernatant was transferred to a fresh tube. Normal mouse IgG (NIgG, as control) or CRBN antibody were added and incubated at 4°C overnight (with rocking). 30 μL of A/G slurry was added and incubated for 1 hour. After washing 3 times with lysis buffer, loading buffer (Cell Signaling Technology) was added and samples were boiled and loaded to 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gels.
Pull-down using Ni+beads. OCI-MY5/vec or OCI-MY5/CRBN-His (untreated or pretreated with lenalidomide for 48 hours) were harvested and stored at –80°C. The cell pellets were resuspended in 1× binding buffer (20 mM Tris/HCl, pH 7.9, 500 mM NaCl, and 10% glycerol) with protease inhibitors and then equilibrated on ice for 30 minutes at 800 psi in a Parr nitrogen cavitation bomb.20 After releasing the pressure, the cell lysates were centrifuged at 33 000g for 30 minutes at 4°C to collect supernatant. Protein amount was quantified and the same amount of proteins from each sample was incubated with Ni-NTA agarose on a rocker for 2 hours at 4°C. The loaded Ni-NTA agarose beads were packed into a column and washed with 1× binding buffer containing 10 mM imidazole (first wash), 20 mM imidazole (second wash), and 40 mM imidazole (third wash).21 The bound proteins were eluted with 1× binding buffer containing 400 mM imidazole and were used for mass spectrometry (MS) analysis.
MS analysis
For proteomics experiments, proteins were separated by 10% SDS-PAGE and were stained with Coomassie blue. Each lane of the SDS-PAGE gel was cut into 10 slices, followed by destaining and digestion with trypsin. The peptides were extracted and then purified by solid-phase extraction (C18 ZipTip; Millipore, Billerica, MA).
High-performance liquid chromatography/electrospray ionization tandem MS (HPLC-ESI-MS/MS) (ThermoScientific, San Jose, CA) was performed on an Orbitrap Elite Velos PRO mass spectrometer fitted with a NanoFlex source (New Objective, Woburn, MA) coupled to a DIONEX (Sunnyvale, CA) Ultimate 3000 Series Nano/Cap System NCS-3500RS. NanoLC was performed using an Acclaim PepMap100 trap column an Acclaim PepMap RSLC analytical column (Thermo Scientific). A “top-10” data-dependent MS/MS analysis was performed. The fragment mass spectra were then searched against the human SwissProt_2013_02 database using Mascot (Matrix Science, London, United Kingdom; version 2.4). Cross-correlation of Mascot search results with X! Tandem was accomplished with Scaffold (version Scaffold_4.2.1; Proteome Software, Portland, OR). Probability assessment of peptide assignments and protein identifications were made through the use of Scaffold. Only peptides with ≥95% probability were considered.
Analysis of IKZF1 expression by flow cytometry
Five hundred thousand cells were resuspended in 0.1 mL phosphate-buffered saline (PBS) in 5-mL U-bottom tubes. Cells were fixed by adding 40 μL 10% formaldehyde at room temperature for 10 minutes and then cooled on ice for 1 minute. 0.6 mL of ice-cold methanol was added and left on ice for 20 to 30 minutes. After washing in PBS buffer, anti-IKZF1 was added and incubated at room temperature for 20 minutes. After washing in PBS buffer, a second Ab was added and incubated at room temperature, kept in the dark for 15 minutes, and washed once in PBS+3% fetal bovine serum, and samples were then analyzed by flow cytometry.
In silico network and pathway enrichment analysis
The Cytoscape Reactome FI plugin was used to assess the molecular function, biological pathways, and additional protein-protein interactions as described.22 Lists of proteins upregulated or downregulated in response to lenalidomide were evaluated against the latest versions of the following databases (covering >50% of human proteins): CellMap, Reactome, KEGG, NCI-PID, and BioCartel. Linker proteins were included in the network modeling, and comparative pathway enrichment predictions were generated using input lists including or excluding linker proteins. Results were filtered for a P value < .05 and a false discovery rate < 0.05.
Primary patient samples
We screened the gene expression profiling (GEP) levels from 44 MM patients from Mayo Clinic homogeneously treated in 2 prospective clinical trials with pomalidomide and dexamethasone.23,24 The first trial included 35 relapsed or refractory patients who received pomalidomide 2 mg daily continuously on a 28-day cycle and dexamethasone 40 mg weekly. The second trial included 35 relapsed or refractory patients who received pomalidomide 4 mg daily continuously on a 28-day cycle and dexamethasone 40 mg weekly. Forty-four of the 70 patients in these 2 trials were successfully analyzed for gene expression before therapy initiation.
GEP
RNA was isolated from marrow CD138+ plasma cells. GEP was performed from total RNA using the Affymetrix U133Plus2.0 array. Microarrays were scanned with an Affymetrix Scanner 3000, and data normalization was performed using Expression Console (Affymetrix) and the robust multi-array average (RMA). RMA intensity values were log2-transformed for subsequent analyses. Public datasets were used for comparative expression studies on different stages of plasma cell neoplasm (GSE6477) and for analyzing the association between copy number and gene expression (http://www.broadinstitute.org/mmgp/home).
Response criteria
Response was assessed according to the International Myeloma Working Group criteria.25 A positive response to therapy in this study required a partial response or better.
Statistical analysis
For the comparison of gene expression levels between groups, we used the Fisher 2-tailed test (comparison between 2 groups) and the Kruskal-Wallis test (≥3 groups).
To evaluate the association of CRBN-binding protein level with progression-free survival (time from registration to the earlier of disease progression or death) and overall survival (OS; time from registration to death), median level was used to separate patients into 2 groups, followed by analysis using the Kaplan-Meier methods. The COX survival model was also used to analyze the effects of 11 proteins identified from CRBN complex altered by lenalidomide on survival outcomes. To evaluate the connection between IKZF1 and response, we determined the quartile cutoff points for IKZF1 expression (for all MM patients). Optimal cutoff points for survival analyses were determined using the Contal and O’Quigley method.
Results
Identification of CRBN interacting proteins
OCI-MY5 MM cells have very low CRBN expression and are highly resistant to IMiDs. We were able to restore IMiD responsiveness by introducing wild-type CRBN with a His-tag at its C-terminal (OCI-MY5 CRBN-His) (supplemental Figure 1, available on the Blood Web site).
We then used an anti-CRBN antibody to immunoprecipitate CRBN and its associated proteins, using a normal mouse IgG-negative control (Figure 1A) and performed quantitative proteomics to define the CRBN interactome. Eighty-three putative CRBN protein–binding partners with an average twofold higher assigned spectra compared with controls were identified from duplicate experiments (supplemental Data 1). The most differentially associated 10 proteins (>fourfold higher than the negative controls in both runs) include CRBN and its known associated proteins, CUL4A and DDB1 (Table 1), confirming the specificity of the method. Six proteins (CRBN, ACAD9, FLOT1, FN1, HDDC3, and IKZF1) were detected in CRBN immunoprecipitates and were completely absent in both negative control immunoprecipitates.
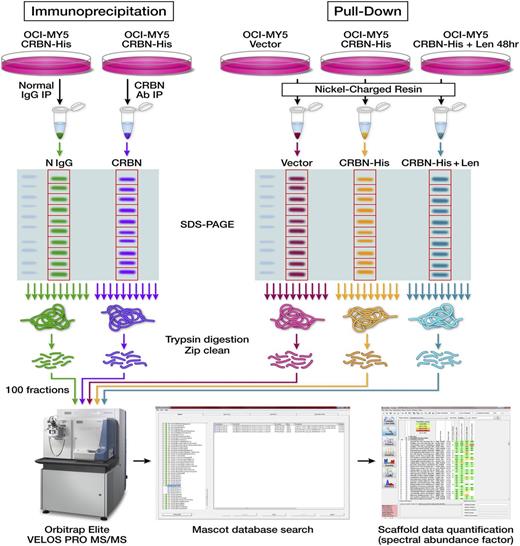
Identification of proteins in CRBN protein-protein interaction. Schematic diagrams show the workflow designed for identifying CRBN-binding partners via Co-IP or Ni+ beads pull-down and proteomic-based analysis. The first approach includes Co-IP, separation by SDS-PAGE, in-gel digestion and peptide extraction, and HPLC-ESI-MS/MS quantitative analysis. The second approach included using Ni+ beads to pull down the proteins from control cells (OCI-MY5 vector) and CRBN-overexpressing cells (OCI-MY5 CRBN-His) untreated or pre-treated with lenalidomide for 48 hours, separation by SDS-PAGE, in-gel digestion and peptide extraction, and HPLC-ESI-MS/MS quantitative analysis.
Identification of proteins in CRBN protein-protein interaction. Schematic diagrams show the workflow designed for identifying CRBN-binding partners via Co-IP or Ni+ beads pull-down and proteomic-based analysis. The first approach includes Co-IP, separation by SDS-PAGE, in-gel digestion and peptide extraction, and HPLC-ESI-MS/MS quantitative analysis. The second approach included using Ni+ beads to pull down the proteins from control cells (OCI-MY5 vector) and CRBN-overexpressing cells (OCI-MY5 CRBN-His) untreated or pre-treated with lenalidomide for 48 hours, separation by SDS-PAGE, in-gel digestion and peptide extraction, and HPLC-ESI-MS/MS quantitative analysis.
Top differentially associated proteins identified in Co-IP
| . | Experiment 1* . | Experiment 2* . | ||
|---|---|---|---|---|
| Proteins . | NlgG . | CRBN Ab . | NlgG . | CRBN Ab . |
| ACAD9 | 0 | 5 | 0 | 4 |
| CRBN | 0 | 3 | 0 | 9 |
| CUL4A | 0 | 2 | 1 | 8 |
| DDB1 | 7 | 123 | 9 | 47 |
| FN1 | 0 | 3 | 0 | 2 |
| FLOT1 | 0 | 2 | 0 | 3 |
| IKZF1 | 0 | 2 | 0 | 2 |
| HDDC3 | 0 | 2 | 0 | 2 |
| EPPK1 | 1 | 4 | 0 | 5 |
| LSM12 | 0 | 3 | 1 | 4 |
| . | Experiment 1* . | Experiment 2* . | ||
|---|---|---|---|---|
| Proteins . | NlgG . | CRBN Ab . | NlgG . | CRBN Ab . |
| ACAD9 | 0 | 5 | 0 | 4 |
| CRBN | 0 | 3 | 0 | 9 |
| CUL4A | 0 | 2 | 1 | 8 |
| DDB1 | 7 | 123 | 9 | 47 |
| FN1 | 0 | 3 | 0 | 2 |
| FLOT1 | 0 | 2 | 0 | 3 |
| IKZF1 | 0 | 2 | 0 | 2 |
| HDDC3 | 0 | 2 | 0 | 2 |
| EPPK1 | 1 | 4 | 0 | 5 |
| LSM12 | 0 | 3 | 1 | 4 |
The number in each column represents the number of assigned spectra for that protein.
We independently also used Ni charged (Ni+) beads to identify CRBN-associated proteins (Figure 1). In duplicate experiments, 171 CRBN-associated proteins with higher assigned spectra (average ≥twofold higher) were identified in the CRBN overexpressing cells vs the negative control vector–expressing cells (supplemental Data 1). The most differentially identified proteins again included CRBN, DDB1, and CUL4A. Over all, 10 proteins were identified to be shared between the Co-IP and Ni+ bead methods, including CRBN, CUL4A, DDB1, IKZF1, KPNA2, LTF, PFKL, PRKAR2A, RANGAP1, and SHMT2 (Table 2). Among the 13 unique proteins (not part of the CRBN E3 ligase complex) shown in Tables 1 and 2, three are involved in metabolic pathways (Phosphofructokinase; ACAD9, a member of the acyl-CoA dehydrogenase family; and PRKAR2A, which engages in lipid and glucose metabolism), and two of these strong CRBN-binding proteins are engaged in nuclear transport (RANGAP1, KPNA2).
The proteins in CRBN complex identified by both Co-IP and pull-down assays
| . | Co-IP . | Ni+ beads pull-down . | ||||||
|---|---|---|---|---|---|---|---|---|
| Experiment 1* . | Experiment 2* . | Experiment 1* . | Experiment 2* . | |||||
| Proteins . | NlgG . | CRBN Ab . | NlgG . | CRBN Ab . | Vector . | CRBN-His . | Vector . | CRBN-His . |
| CRBN | 0 | 3 | 0 | 9 | 0 | 32 | 0 | 35 |
| CUL4A | 0 | 2 | 1 | 8 | 0 | 34 | 1 | 31 |
| DDB1 | 7 | 123 | 9 | 47 | 11 | 122 | 3 | 82 |
| IKZF1 | 0 | 2 | 0 | 2 | 2 | 3 | 0 | 6 |
| KPNA2 | 8 | 14 | 1 | 8 | 5 | 36 | 0 | 16 |
| LTF | 2 | 5 | 1 | 4 | 3 | 16 | 1 | 33 |
| PFKL | 1 | 4 | 2 | 4 | 5 | 12 | 0 | 3 |
| PRKAR2A | 4 | 9 | 1 | 2 | 7 | 11 | 1 | 6 |
| RANGAP1 | 3 | 9 | 2 | 4 | 3 | 5 | 0 | 5 |
| SHMT2 | 10 | 25 | 3 | 8 | 4 | 8 | 1 | 4 |
| . | Co-IP . | Ni+ beads pull-down . | ||||||
|---|---|---|---|---|---|---|---|---|
| Experiment 1* . | Experiment 2* . | Experiment 1* . | Experiment 2* . | |||||
| Proteins . | NlgG . | CRBN Ab . | NlgG . | CRBN Ab . | Vector . | CRBN-His . | Vector . | CRBN-His . |
| CRBN | 0 | 3 | 0 | 9 | 0 | 32 | 0 | 35 |
| CUL4A | 0 | 2 | 1 | 8 | 0 | 34 | 1 | 31 |
| DDB1 | 7 | 123 | 9 | 47 | 11 | 122 | 3 | 82 |
| IKZF1 | 0 | 2 | 0 | 2 | 2 | 3 | 0 | 6 |
| KPNA2 | 8 | 14 | 1 | 8 | 5 | 36 | 0 | 16 |
| LTF | 2 | 5 | 1 | 4 | 3 | 16 | 1 | 33 |
| PFKL | 1 | 4 | 2 | 4 | 5 | 12 | 0 | 3 |
| PRKAR2A | 4 | 9 | 1 | 2 | 7 | 11 | 1 | 6 |
| RANGAP1 | 3 | 9 | 2 | 4 | 3 | 5 | 0 | 5 |
| SHMT2 | 10 | 25 | 3 | 8 | 4 | 8 | 1 | 4 |
The number in each column represents the number of assigned spectra for that protein.
CRBN-interacting proteins relevant to IMiD treatment
We next examined the influence of lenalidomide treatment. Proteomic analysis identified 114 proteins that decreased in CRBN-binding abundance (≥twofold lower in the Ni+ beads pull-down samples from OCI-MY5/CRBN cells 48 hours after lenalidomide treatment vs no treatment, n = 3, supplemental Data 1). Of these 114 proteins, 46 were also identified to significantly bind to CRBN complex in the absence of drug. Two of the most downregulated CRBN-associated proteins after lenalidomide treatment included the Ikaros family members IKZF1 (Ikaros) and IKZF3 (Aiolos) (Table 3). Alternatively, 16 proteins underwent an increase in abundance after lenalidomide treatment (supplemental Data 1), including CRBN and its associated proteins, DDB1, CUL4B, CUL4A, and SQSTM1 (Table 3).
Top lenalidomide-regulated putative CRBN-interacting proteins
| . | Experiment 1* . | Experiment 2* . | Experiment 3* . | . | |||
|---|---|---|---|---|---|---|---|
| Proteins . | –Len . | +Len . | –Len . | +Len . | –Len . | +Len . | Regulation . |
| GNL2 | 2 | 0 | 2 | 0 | 2 | 0 | Down |
| STUB1 | 2 | 0 | 2 | 0 | 2 | 0 | Down |
| IKZF1 | 5 | 0 | 6 | 0 | 3 | 0 | Down |
| IKZF3 | 5 | 0 | 7 | 0 | 10 | 0 | Down |
| ANKRD12 | 2 | 1 | 4 | 0 | 6 | 0 | Down |
| KPNA2 | 15 | 2 | 16 | 9 | 36 | 19 | Down |
| CUL4A | 30 | 66 | 34 | 45 | 31 | 58 | Up |
| CUL4B | 18 | 43 | 9 | 45 | 44 | 130 | Up |
| DDB1 | 112 | 179 | 82 | 261 | 122 | 271 | Up |
| SQSTM1 | 11 | 17 | 7 | 17 | 12 | 21 | Up |
| . | Experiment 1* . | Experiment 2* . | Experiment 3* . | . | |||
|---|---|---|---|---|---|---|---|
| Proteins . | –Len . | +Len . | –Len . | +Len . | –Len . | +Len . | Regulation . |
| GNL2 | 2 | 0 | 2 | 0 | 2 | 0 | Down |
| STUB1 | 2 | 0 | 2 | 0 | 2 | 0 | Down |
| IKZF1 | 5 | 0 | 6 | 0 | 3 | 0 | Down |
| IKZF3 | 5 | 0 | 7 | 0 | 10 | 0 | Down |
| ANKRD12 | 2 | 1 | 4 | 0 | 6 | 0 | Down |
| KPNA2 | 15 | 2 | 16 | 9 | 36 | 19 | Down |
| CUL4A | 30 | 66 | 34 | 45 | 31 | 58 | Up |
| CUL4B | 18 | 43 | 9 | 45 | 44 | 130 | Up |
| DDB1 | 112 | 179 | 82 | 261 | 122 | 271 | Up |
| SQSTM1 | 11 | 17 | 7 | 17 | 12 | 21 | Up |
The number in each column represents the number of assigned spectra for that protein.
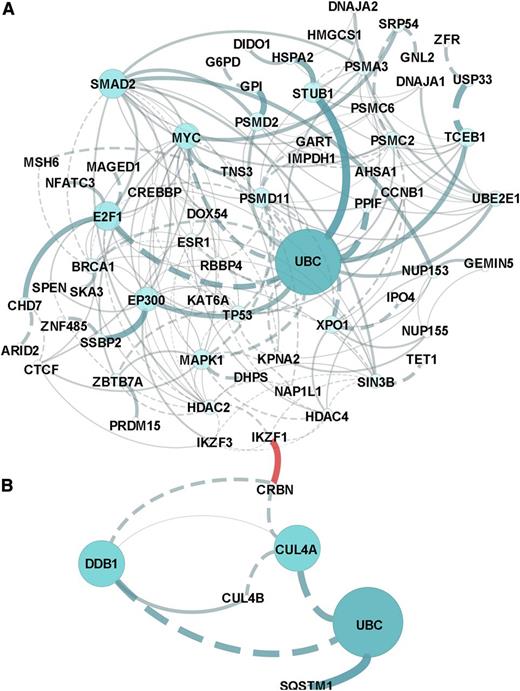
Pathway analysis of all CRBN-binding proteins altered by lenalidomide revealed that the molecular pathways most affected included the regulation of cytoplasmic and nucleus SMAD2/3 signaling; ubiquitin-mediated proteolysis; P53 tumor-suppressor pathways; and signaling by HDAC, NOTCH, and Wnt (Figure 2 and supplemental Data 2). Prominent networks implicated the transcriptional co-activator and histone deacteylase EP300, the proteasome and ubiquitin complex, and MYC complex–associated proteins E2F1 and XPO1.
In silico network and pathway enrichment analysis. Lists of the proteins in the CRBN complex that upregulated (A) or downregulated (B) in response to lenolidomide were evaluated against the latest versions of the following databases (covering >50% of human proteins): CellMap, Reactome, KEGG, NCI-PID, and BioCartel. Linker genes were included in the network modeling and pathway enrichment predictions. Results were filtered for a P value < .05 and a false discovery rate < 0.05. The separate networks modeled from downregulation vs upregulation lists are presented in (A) and (B), and the association or connection between IKZF1 and CRBN was a custom annotation added to the figures.
In silico network and pathway enrichment analysis. Lists of the proteins in the CRBN complex that upregulated (A) or downregulated (B) in response to lenolidomide were evaluated against the latest versions of the following databases (covering >50% of human proteins): CellMap, Reactome, KEGG, NCI-PID, and BioCartel. Linker genes were included in the network modeling and pathway enrichment predictions. Results were filtered for a P value < .05 and a false discovery rate < 0.05. The separate networks modeled from downregulation vs upregulation lists are presented in (A) and (B), and the association or connection between IKZF1 and CRBN was a custom annotation added to the figures.
Validation of IKZF1 and IKZF3 as CRBN-binding proteins
Immunoblotting (Figure 3A) confirmed that CRBN, DDB1, and CUL4A were enriched in the CRBN binding fraction from OCI-MY5/CRBN-His but not from control cells (vector). After lenalidomide treatment, CRBN, DDB1, and CUL4A were also more abundant in the CRBN pull-down complex, whereas IKZF1, IKZF3, and KPNA2 decreased (Figure 3A). We found that both IKZF1 and IKZF3 were downregulated within 3 to 6 hours of lenalidomide treatment and that this change occurred significantly before downregulation of IRF4, MYC, and other binding proteins such as KPNA2 (Figure 3B-C).
Validation of IKZF1 and IKZF3 as CRBN-binding proteins. (A) Immunoblotting of the Ni+ beads pull-down proteins from OCI-MY5 vector (control) and OCI-MY5 CRBN-His with (+) without (–) lenalidomide treatment of 48 hours (2 independent experiments). CRBN, DDB1, CUL4A, IKZF1, IKZF3, and KPNA2 showed similar changes detected by MS analysis. (B-C) OCI-MY5 CRBN-His and MM1.S cells were treated with lenalidomide (10 μM) in a time course, and the changes of some top proteins identified from MS analysis or related with lenalidomide-induced cytotoxicity were measured by western blot. IKZF1 and IKZF3 downregulation was identified as the earliest change after lenalidomide treatment. (D) OCI-MY5 CRBN-His cells were treated with dimethyl sulfoxide or lenalidomide for 3 hours, and Co-IP was performed with anti-CRBN antibody to immunoprecipitate CRBN and CRBN-associated proteins, using a normal mouse IgG as a negative control. IKZF1 and IKZF3 were greatly increased in CRBN antibody–immunoprecipitated samples after lenalidomide treatment (left panels), but they are decreased in cell lysate after lenalidomide treatment (right panels). CRBN increased in both immunoprecipitated and nonimmunoprecipitated lysate.
Validation of IKZF1 and IKZF3 as CRBN-binding proteins. (A) Immunoblotting of the Ni+ beads pull-down proteins from OCI-MY5 vector (control) and OCI-MY5 CRBN-His with (+) without (–) lenalidomide treatment of 48 hours (2 independent experiments). CRBN, DDB1, CUL4A, IKZF1, IKZF3, and KPNA2 showed similar changes detected by MS analysis. (B-C) OCI-MY5 CRBN-His and MM1.S cells were treated with lenalidomide (10 μM) in a time course, and the changes of some top proteins identified from MS analysis or related with lenalidomide-induced cytotoxicity were measured by western blot. IKZF1 and IKZF3 downregulation was identified as the earliest change after lenalidomide treatment. (D) OCI-MY5 CRBN-His cells were treated with dimethyl sulfoxide or lenalidomide for 3 hours, and Co-IP was performed with anti-CRBN antibody to immunoprecipitate CRBN and CRBN-associated proteins, using a normal mouse IgG as a negative control. IKZF1 and IKZF3 were greatly increased in CRBN antibody–immunoprecipitated samples after lenalidomide treatment (left panels), but they are decreased in cell lysate after lenalidomide treatment (right panels). CRBN increased in both immunoprecipitated and nonimmunoprecipitated lysate.
Binding of IKZF1 and IKZF3 to CRBN was actually initially increased after lenalidomide treatment (3 hours), whereas protein levels in lysates were decreased (Figure 3D), suggesting a rapid binding and then later degradation of IKZF1 and IKZF3. The CRBN level in Co-IP samples increased after lenalidomide treatment, but unlike Ikaros proteins, the CRBN protein level increased in lysates (Figure 3D).
Drug resistance and CRBN target proteins in MM
We compared Ikaros and Aiolos (IKZF1 and IKZF3) gene expression levels across different human tissues and malignancies. IKZF1 (Figure 4A) and IKZF3 (Figure 4B) were expressed in most hematological malignancies, including MM, but not in solid tumors (source: Broad-Novartis Encyclopedia and The Genotype Tissue Expression Portal).26,27
Gene expression analysis of IKZF1 and IKZF3 across cell lines originated from multiple hematological and solid tumors. The number of cases of each tumor entity is indicated in parenthesis. Red boxes indicate the set of MM cell lines. Results show IKZF1 (A) and IKZF3 (B) expression in most cell lines established from hematological tumors. The findings in hematological neoplasms contrast with the lack of expression of both genes found in solid-tumor cell lines. AML, acute myeloid leukemia; B-ALL, B-cell acute lymphoblastic leukemia; CML, chronic myeloid leukemia; T-ALL, T-cell acute lymphoblastic leukemia; MCL, mantle cell lymphoma; DLBCL, diffuse large B-cell lymphoma; MM, multiple myeloma.26,27
Gene expression analysis of IKZF1 and IKZF3 across cell lines originated from multiple hematological and solid tumors. The number of cases of each tumor entity is indicated in parenthesis. Red boxes indicate the set of MM cell lines. Results show IKZF1 (A) and IKZF3 (B) expression in most cell lines established from hematological tumors. The findings in hematological neoplasms contrast with the lack of expression of both genes found in solid-tumor cell lines. AML, acute myeloid leukemia; B-ALL, B-cell acute lymphoblastic leukemia; CML, chronic myeloid leukemia; T-ALL, T-cell acute lymphoblastic leukemia; MCL, mantle cell lymphoma; DLBCL, diffuse large B-cell lymphoma; MM, multiple myeloma.26,27
IKZF1 and IKZF3 gene expression levels did not change in different stages of disease progression from normal polyclonal to monoclonal gammopathy of undetermined significance, smoldering multiple myeloma, and MM. Furthermore, no significant differences were found in the expression level across genetic subtypes (data not shown).
Western blot analysis revealed that IKZF1 levels are adequate in all sensitive MM lines examined, whereas 3 of 5 tested lenalidomide-resistant cell lines IKZF1 were low (Figure 5A). As an alternate and potentially clinically useful strategy, we were also able to detect IKZF1 expression and its changes after IMiD using flow cytometry analysis, and the result was consistent with the western blot results (Figure 5B).
Expression of Ikaros proteins in HMCLs and induction of cytotoxicity after knockdown of IKZF1 and IKZF3. (A) Expression of IKZF1 and IKZF3 in HMCLs. Nine HMLCs either sensitive or resistant to lenalidomide were analyzed for the expression of Ikaros by western blot. (B) Expression of IKZF1 in HMCLs was detected by flow cytometry analysis. (C-D) Knockdown of IKZF1 and IKZF3 induced cytotoxicity. MM1.S cells were infected with lentivirus-expressing nontargeting control or IKZF1 or IKZF3 shRNAs. MTT assays were performed at day 6 after infection and western blot analysis was performed at day 3 after infection. (E-G) Overexpression of IKZF1 and IKZF3 in a lenalidomide-sensitive cell line, MM1.S, did not substantially change lenalidomide sensitivity. MM1.S cells were infected with control virus (expressing vector alone) or lentivirus-expressing IKZF1 and IKZF3 after 72 hours (E), or after sorting GFP+ cells in MM1.S cells infected by IKZF1 lentivirus at 14 days after infection (F), cells were seeded in a 96-well plate and were treated with lenalidomide. (G) MTT assay was performed at day 6 after treatment and western blot analysis was performed at day 3 to confirm overexpression of IKZF1 and IKZF3.
Expression of Ikaros proteins in HMCLs and induction of cytotoxicity after knockdown of IKZF1 and IKZF3. (A) Expression of IKZF1 and IKZF3 in HMCLs. Nine HMLCs either sensitive or resistant to lenalidomide were analyzed for the expression of Ikaros by western blot. (B) Expression of IKZF1 in HMCLs was detected by flow cytometry analysis. (C-D) Knockdown of IKZF1 and IKZF3 induced cytotoxicity. MM1.S cells were infected with lentivirus-expressing nontargeting control or IKZF1 or IKZF3 shRNAs. MTT assays were performed at day 6 after infection and western blot analysis was performed at day 3 after infection. (E-G) Overexpression of IKZF1 and IKZF3 in a lenalidomide-sensitive cell line, MM1.S, did not substantially change lenalidomide sensitivity. MM1.S cells were infected with control virus (expressing vector alone) or lentivirus-expressing IKZF1 and IKZF3 after 72 hours (E), or after sorting GFP+ cells in MM1.S cells infected by IKZF1 lentivirus at 14 days after infection (F), cells were seeded in a 96-well plate and were treated with lenalidomide. (G) MTT assay was performed at day 6 after treatment and western blot analysis was performed at day 3 to confirm overexpression of IKZF1 and IKZF3.
Using lentiviral shRNA expression, knockdown of IKZF1 and IKZF3 induced a reduction of MM cell viability and downregulation of IRF4 (Figure 5C-D); indeed, no stable MM line with low IKZF1 could be generated by this method, although MM cell lines with low IKZF1 expression do exist. Forced overexpression of IKZF1 and IKZF3 did decrease lenalidomide sensitivity, albeit not dramatically (Figure 5E-G).
To explain the low IKZF1 in cell lines, we next screened public data sets and 69 MM cell lines for mutations in IKZF1 and IKZF3. As with CRBN, mutations were very uncommon in primary MM, with <1% of MM patients harboring nonsynonymous mutations in either IKZF1 or IKZF3. It is important to note, however, that these sequence data are generated from patients who are mostly early in their treatment course. In contrast, mutations of IKZF1/3 were observed in 6 of 69 MM cell lines (8.7%) (supplemental Data 3), including an LR208R single amino acid deletion resulting from an in-frame L208R substitution, resulting in a codon deletion in the FR4 cell line, which has very low levels of IKZF1 and which is highly IMiD-resistant, likely also the result of high IRF4 levels from a known translocation t(6;14) IgH-IRF4. Recurrent mutations at Q156K in both the XG1 and H1112 cell lines and at G158R in OCI-MY7 all occur within the critical amino acid–binding domain of IKZF1 and IKZF3 to CRBN. The significance of the Q156K mutation is unknown because XG1 cells are very sensitive to IMiD therapy. Further evidence of pathway mutation is evident in the observation that IRF4 mutations are found in 4% of MM genomes sequenced and the t(6;14) IgH-IRF4 is present in 3 of 69 cell lines. Because mutations or deletions of CRBN have also been reported in drug-resistant patients, it seems that mutation of the CRBN-IKZF-IRF4 pathway may end up explaining IMiD resistance in more patients than was previously suspected.
Correlation of CRBN-binding proteins with IMiD sensitivity and survival outcomes
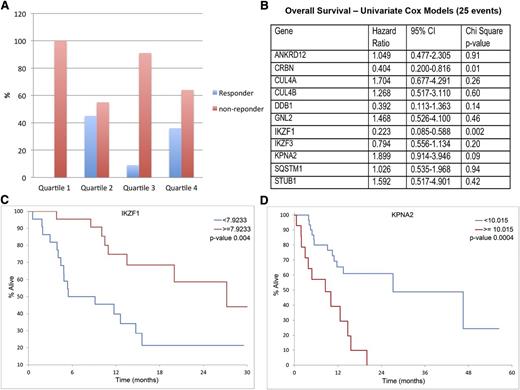
Next, we used GEP to examine the association between drug response and survival for the 11 top lenalidomide-regulated CRBN-interacting proteins. Baseline gene expression levels were measured in a cohort of 44 refractory MM patients before initiation of pomalidomide and dexamethasone therapy as part of phase 2 clinical trials.23,24 Responders were defined as patients achieving partial response or higher after 2 cycles of treatment. No significant difference was observed in responders and nonresponders between the high and low IKZF1 expression groups. However, when we set the quartile cutoff points for IKZF1 expression (for all MM patients), we found a lack of clinical response for patients in the lowest quartile range of IKZF1 expression. The response rate was 0% in the lowest quartile of IKZF1 expression vs 30% in the highest 3 quartiles (P = .04) (Figure 6A). Conversely, IKZF3, Eos (IKZF4), and Pegasus (IKZF5) did not show differences between responders and nonresponders to pomalidomide.
Correlation of IMiD responsiveness and OS with IKZF1 and KPNA2 gene expression level in pomalidomide and dexamethasone therapy. A cohort of 44 MM patients treated with pomalidomide with GEP measured before initiation of chemotherapy was used in this study. (A) Responsiveness to pomalidomide in quartiles of IKZF1 expression. No patient in the lowest IKZF1 quartile expression responded to pomalidomide treatment. (B)The gene expression of the 11 top lenalidomide-regulated putative CRBN-interacting proteins was analyzed to examine the association between drug response and survival by using COX modeling. IKZF1 was identified as the top gene, which expression is associated with OS (P = .002). (C-D) Low IKZF1 and high KPNA2 expression levels were correlated with shorter OS.
Correlation of IMiD responsiveness and OS with IKZF1 and KPNA2 gene expression level in pomalidomide and dexamethasone therapy. A cohort of 44 MM patients treated with pomalidomide with GEP measured before initiation of chemotherapy was used in this study. (A) Responsiveness to pomalidomide in quartiles of IKZF1 expression. No patient in the lowest IKZF1 quartile expression responded to pomalidomide treatment. (B)The gene expression of the 11 top lenalidomide-regulated putative CRBN-interacting proteins was analyzed to examine the association between drug response and survival by using COX modeling. IKZF1 was identified as the top gene, which expression is associated with OS (P = .002). (C-D) Low IKZF1 and high KPNA2 expression levels were correlated with shorter OS.
Expression of CRBN, IKZF1, and KPNA2 correlated with OS after this therapy (Figure 6B). Patients with low IKZF1 expression had a median OS of 7.3 months compared with 27.2 months in the patients with higher IKZF1 expression (P = .004) (Figure 6C). The group with lower IKZF1 expression also exhibited shorter median progression-free survival (4.9 months) compared with the patients with a higher IKZF1 expression (7.3 months), but the differences were not statistically significant. In addition to IKZF1 and CRBN (previously published), high KPNA2 expression level was also found to be associated with shorter OS (Figure 6D).
We then analyzed the Apex clinical trial data set,28 a trial that compared MM patients treated with single-agent dexamethasone or bortezomib. No significant difference in the response rate between patients with the lowest and the highest quartile of IKZF1 expression was found (data not shown). We noted that IKZF1 low expressors have a longer OS with bortezomib (supplemental Figure 2A), the opposite of results from the pomalidomide trial. We also demonstrated that lenalidomide-resistant cell lines with low IKZF1 expression are just as responsive to bortezomib as lenalidomide-sensitive MM cell lines with high IKZF1 expression (supplemental Figure 2A)
Discussion
In this study, we sought to determine CRBN-binding proteins and to establish their relevance to MM biology by exploring changes in protein abundance after exposure to the IMiD lenalidomide. Two independent pull-down methods were used to isolate proteins bound in the CRBN complex, followed by identification via MS analysis. Both methods identified CRBN and its known associated proteins, DDB1 and CUL4A, as among the highest associating proteins, demonstrating the validity of methodologies. Ten proteins were identified in the CRBN complex by both methods, including IKZF1, a protein that has recently been identified by others as a substrate of CRBN E3 ubiquitin ligase activity.17,18 We also identified the second implicated Ikaros member, IKZF3, as one of the CRBN-binding proteins most downregulated after lenalidomide treatment of 48 hours. IKZF1 and IKZF3 are members of the Ikaros zinc-finger proteins, and both are chromatin-remodeling transcription regulators involved in the regulation of lymphocyte development.29-33 IKZF1 and IKZF3 were previously demonstrated to regulate B-cell activation and differentiation.30 Dysregulated IKZF1 and IKZF3 are associated with B-cell acute lymphoblastic leukemia and B-cell chronic lymphocytic leukemia.34-37 Recently, 2 landmark studies have independently shown that IKZF1 and IKZF3 are substrates of CRBN ubiquitin ligase.17,18 Consistent with these studies, we validated that binding of IKZF1 and IKZF3 with CRBN was greatly enhanced within a few hours of lenalidomide treatment, but that IKZF1 and IKZF3 levels are later downregulated in the same samples, suggesting they are substrates of CRBN ubiquitin ligase. However, our data also identified that CRBN and DDB1, in contrast to Ikaros proteins, are upregulated in the same pull-down, implying their degradation is blocked after lenalidomide treatment. This result was consistent with previous studies that suggest that CRBN itself is a substrate of CRBN ubiquitin ligase. A possible explanation for this paradox is that after binding to IMiDs, CRBN may become resistant to proteasomal degradation. Theoretically, accumulated CRBN will enhance its function.
Consistent with recent reports, knockdown of IFZF1 and IKZF3 in our study induced MM cytotoxicity, confirming that this family is important for MM survival. Interestingly, although IKZF1 and IKZF3 were downregulated within hours after lenalidomide treatment in most MM cell lines, it usually takes another 24 to 72 hours to see lenalidomide-induced downregulation of IRF4 and associated MM cytotoxicity, suggesting that additional interactions or signaling cascades may be required to achieve a full drug response. We have observed but cannot yet explain why some lenalidomide-resistant cell lines have downregulation of IKZF1 and IKZF3 after lenalidomide treatment, but do not have lenalidomide-induced downregulation of IRF4 and cytotoxicity (data not shown). Another unexplained observation we have made is that the doses required to downregulate Ikaros are also much lower than those required to mediate MM cytotoxicity.
In addition to Ikaros family members, our MS data identified other interesting proteins that may bind the CRBN complex and are downregulated by IMiDs, including several binding partners involved in metabolism, nuclear transport, and protein folding such as KPNA2, NUP153, and DNAJA2. Interestingly, several proteasome subunits were also detected, consistent with a recent report that CRBN was identified by two-hybrid screen to associate with a proteasome subunit.38 The significance of this observation is not yet understood but is provocative because the scientific evidence would currently suggest that proteasome inhibitors and IMiDs work in opposite directions in either, requiring proteasomal degradation or inhibiting this function, a scientific observation at odds with the clinical experience.
KPNA2 is noted because this nuclear transport protein has also been associated with B-cell development via Pax5.39 KPNA2 appears to be depleted later than the Ikaros family. Our data suggest that low levels of KPNA2 at baseline (possibly as a marker for likelihood of drug activity) are associated with a better prognosis.
We found in the current study that low IKZF1 expression levels are associated with poor response to IMiDs and shorter OS. The response rate was 0% in the lowest quartile of IKZF1 expression vs 30% in the highest 3 quartiles (P = .04). This observation appears to contradict our data and those of recent publications that deliberate that overexpression of IKZF1 in MM cell lines induced resistance to lenalidomide.17,18 A possible explanation is that myeloma cells surviving with low IKZF1 are less dependent on IKZF1-associated signaling for proliferation and survival and are therefore resistant to IMiDs. In at least one lenalidomide-resistant cell line, FR4, a genomic explanation of resistance emerges in that FR4 has a predicted damaging genomic mutation of IKZF1, very low levels of IKZF1 expression, and a translocation upregulating IRF4, which together would seem to explain the ability to survive without IKZF1.
In summary, this study demonstrates numerous novel CRBN-binding proteins altered in MM cells after IMiD treatment and confirms recent work implicating the Ikaros transcription factors as important members of this group. Unique to our study is not only the identification of novel CRBN-binding proteins but also the correlation of IKZF1 and KPNA2 with clinical outcome of IMiD therapy.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported by a National Cancer Institute grant (CA 183968) and a Leukemia and Lymphoma Society grant (LLS-7A).
Authorship
Contribution: A.K.S., Y.X.Z., E.B., X.B., P.L.B., and R.F. contributed to conception and design of the study; Y.X.Z., E.B., C.X.S., L.B., J.S., K.M.K., P.L., M.L., P.J., B.L., K.L., J.M., M.L., and M.D.C. contributed to acquisition of data and analysis of the data; all authors contributed to interpretation of data; Y.X.Z., E.B., and A.K.S. drafted the article; and all authors read, revised the article critically for important intellectual content, and gave final approval of the version to be published.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: A. Keith Stewart, Division of Hematology, Mayo Clinic Arizona, 13400 E Shea Blvd, Scottsdale, AZ 85259; e-mail: stewart.keith@mayo.edu.