Key Points
The expression level of patient HLA-C allotypes affects GVHD and mortality after HCT from HLA-C-mismatched unrelated donors.
Transplant outcome can be improved by avoiding high-risk HLA-C-mismatched donors when no matched stem cell source is available.
Abstract
Life-threatening graft-versus-host disease (GVHD) limits the use of HLA-C-mismatched unrelated donors in transplantation. Clinicians lack criteria for donor selection when HLA-C-mismatched donors are a patient’s only option for cure. We examined the role for HLA-C expression levels to identify permissible HLA-C mismatches. The median fluorescence intensity, a proxy of HLA-C expression, was assigned to each HLA-C allotype in 1975 patients and their HLA-C-mismatched unrelated transplant donors. The association of outcome with the level of expression of patients’ and donors’ HLA-C allotypes was evaluated in multivariable models. Increasing expression level of the patient’s mismatched HLA-C allotype was associated with increased risks of grades III to IV acute GVHD, nonrelapse mortality, and mortality. Increasing expression level among HLA-C mismatches with residue 116 or residue 77/80 mismatching was associated with increased nonrelapse mortality. The immunogenicity of HLA-C mismatches in unrelated donor transplantation is influenced by the expression level of the patient’s mismatched HLA-C allotype. HLA-C expression levels provide new information on mismatches that should be avoided and extend understanding of HLA-C-mediated immune responses in human disease.
Introduction
The transplantation barrier is defined by the HLA genes that are responsible for tissue histocompatibility.1-7 Mismatching for HLA-C allotypes between patients and unrelated donors generally leads to very high risks of acute graft-versus-host disease (GVHD) and mortality after hematopoietic cell transplantation, although risks to individual patients may vary.3-7 The success of transplantation for a given patient may depend on the unique features of the HLA-C mismatch itself. Three different models of HLA-C mismatching shed light on the variability of individual risks. Mismatching can occur between allotypes that elicit an antibody (serologic) response (antigen mismatches) or between allotypes that differ for limited nucleotide sequence variation (allele mismatches). The similarity of sequence features between allele mismatches may contribute to their lower immunogenicity.3-7
A second model of HLA-C alloreactivity entails mismatching for amino acid residues that determine the repertoire of peptides presented to T cells. Patient-donor differences at several residues of the class I molecule might significantly affect the immunogenicity of HLA-C mismatches, and of these residues, residue 116 in the F pocket of the peptide binding groove has a high frequency of patient-donor mismatching and consistently shows an effect on transplant outcome.8-10 HLA-C-mismatched patients who are residue 116 mismatched have higher risks of acute GVHD and mortality than HLA-C-matched patients,10-12 observations that support a critical role for T-cell recognition of class I-peptide complexes.13,14 Most recently, a third model has been proposed in which transplant outcome may depend on the regulation of donor natural killer (NK) cell responses against patient cells.15 Amino acid substitutions at HLA-C residues 77 and 80 define 2 mutually exclusive groups of ligands, each recognized by different killer immunoglobulin-like receptors (KIRs). HLA-C-mismatched patients who are residues 77/80 mismatched may have different transplant outcomes than HLA-C-mismatched patients who are residues 77/80 matched.15-20
Each of the 3 mismatch models suggests that some HLA-C mismatches are less risky than others and therefore represent mismatches that could be considered when matched donors are not available.21 The high overall risks associated with transplantation of HLA-C-mismatched unrelated donors have led some clinicians to abandon the use of such donors altogether. Clinical practice is heterogeneous because the features that define permissive HLA-C mismatches remain ill defined.
Recently, the range of expression across HLA-C allotypes has been elucidated.22 Each serologically defined HLA-C allotype has a characteristic median fluorescence intensity (MFI) of cell surface expression that is reproducible in both healthy and HIV-infected cells in vitro.22 The MFI coefficient is superior to any other marker of expression level, including the previously described single nucleotide polymorphism that resides 35 kb upstream of HLA-C,23 because the MFI provides direct allotype-specific measurement of HLA-C surface expression. Expected levels of HLA-C cell surface expression based on the sum of the 2 allelic MFI coefficients was shown to predict observed HLA-C expression levels among individuals in 2 cohorts, indicating that MFI coefficients can be assigned to each HLA-C allotype in lieu of direct ascertainment of expression.22 Thus, the clinical importance of HLA-C expression can be determined in large-scale retrospective outcome studies where appropriate materials for measuring HLA-C expression directly are not available. Using this approach, higher MFI levels were shown to correlate with better control of HIV viral load and slower progression to HIV-AIDS across ethnic groups, but with increased susceptibility to Crohn disease,22 solidifying the role for HLA-C expression levels in modulating the strength of immune responses. Accordingly, we applied the MFI as a quantitative proxy of HLA-C expression level (simply termed as expression level throughout the manuscript) to assess the clinical significance of the level of HLA-C expression in an exceptionally large international population of patients and unrelated transplant donors whose only HLA mismatch was a single HLA-C allotype.
Materials and methods
Study population, HLA, and MFI
To test the hypothesis that the permissivity of HLA-C mismatches depends on the level of expression of the patient’s and donor’s mismatched HLA-C allotypes, we retrospectively analyzed 1975 patients who received a hematopoietic cell transplant from an HLA-A, -B, -DRB1, and -DQB1 matched but single HLA-C-mismatched unrelated donor as previously defined (Table 1).6,7,24,25 Restriction of the study population to pairs with only 1 HLA-C mismatch removes any contribution of disparity at other HLA loci and addresses whether, among the spectrum of HLA-C mismatches, there are combinations of mismatches that are better tolerated than others.
Demographics of the study population*
| Characteristic . | Transplants (N = 1975) . |
|---|---|
| Patient age, median years (range) | 36.8 (0.19-72.4) |
| Donor age, median years (range) | 36.1 (18-61.1) |
| Year of transplantation | |
| 1983-1992 | 74 (4%) |
| 1993-1999 | 548 (28%) |
| 2000-2005 | 880 (45%) |
| 2006-2011 | 436 (22%) |
| Unknown | 37 (2%) |
| Patient-donor sex | |
| Male-male | 762 (39%) |
| Male-female | 397 (20%) |
| Female-male | 433 (22%) |
| Female-female | 366 (19%) |
| Unknown | 17 (<1%) |
| Disease/early, intermediate, late or advanced, other, or unknown† | |
| Acute leukemia | 974 (49%)/325, 347, 251, 51 |
| Chronic myeloid leukemia | 374 (19%)/237, 95, 21, 21 |
| Myelodysplastic syndrome | 241 (12%)/45, 0, 99, 97 |
| Lymphoma | 192 (10%)/ 6, 3, 45, 138 |
| Multiple myeloma | 27 (1%)/ 1, 0, 18, 8 |
| Other malignancies | 12 (<1%), NA |
| Nonmalignancies | 155 (8%), NA |
| Transplant type | |
| Myeloablative/no TBI | 409 (21%) |
| Myeloablative/ TBI | 967 (49%) |
| Reduced-intensity/nonmyeloablative | 482 (24%) |
| Unknown | 117 (6%) |
| Source of cells | |
| Bone marrow | 1157 (59%) |
| eripheral blood stem cells | 779 (39%) |
| Unknown | 39 (2%) |
| GVHD prophylaxis | |
| Any single agent by itself | 42 (2%) |
| Two or more agents mixed together | 1004 (51%) |
| T-cell depletion | 798 (40%) |
| Other combinations | 29 (2%) |
| Missing | 102 (5%) |
| Allele and antigen mismatches | |
| Allele | 389 (20%) |
| Antigen | 1582 (80%) |
| Unknown‡ | 4 (<15) |
| Residue 116 status of the nonshared patient-donor allotypes | |
| Matched | 847 (43%) |
| Mismatched | 1108 (56%) |
| Unknown | 20 (<1%) |
| Residue 77/80 status of the nonshared patient-donor allotypes | |
| Matched | 955 (48%) |
| Mismatched | 996 (50%) |
| Unknown | 24 (1%) |
| Characteristic . | Transplants (N = 1975) . |
|---|---|
| Patient age, median years (range) | 36.8 (0.19-72.4) |
| Donor age, median years (range) | 36.1 (18-61.1) |
| Year of transplantation | |
| 1983-1992 | 74 (4%) |
| 1993-1999 | 548 (28%) |
| 2000-2005 | 880 (45%) |
| 2006-2011 | 436 (22%) |
| Unknown | 37 (2%) |
| Patient-donor sex | |
| Male-male | 762 (39%) |
| Male-female | 397 (20%) |
| Female-male | 433 (22%) |
| Female-female | 366 (19%) |
| Unknown | 17 (<1%) |
| Disease/early, intermediate, late or advanced, other, or unknown† | |
| Acute leukemia | 974 (49%)/325, 347, 251, 51 |
| Chronic myeloid leukemia | 374 (19%)/237, 95, 21, 21 |
| Myelodysplastic syndrome | 241 (12%)/45, 0, 99, 97 |
| Lymphoma | 192 (10%)/ 6, 3, 45, 138 |
| Multiple myeloma | 27 (1%)/ 1, 0, 18, 8 |
| Other malignancies | 12 (<1%), NA |
| Nonmalignancies | 155 (8%), NA |
| Transplant type | |
| Myeloablative/no TBI | 409 (21%) |
| Myeloablative/ TBI | 967 (49%) |
| Reduced-intensity/nonmyeloablative | 482 (24%) |
| Unknown | 117 (6%) |
| Source of cells | |
| Bone marrow | 1157 (59%) |
| eripheral blood stem cells | 779 (39%) |
| Unknown | 39 (2%) |
| GVHD prophylaxis | |
| Any single agent by itself | 42 (2%) |
| Two or more agents mixed together | 1004 (51%) |
| T-cell depletion | 798 (40%) |
| Other combinations | 29 (2%) |
| Missing | 102 (5%) |
| Allele and antigen mismatches | |
| Allele | 389 (20%) |
| Antigen | 1582 (80%) |
| Unknown‡ | 4 (<15) |
| Residue 116 status of the nonshared patient-donor allotypes | |
| Matched | 847 (43%) |
| Mismatched | 1108 (56%) |
| Unknown | 20 (<1%) |
| Residue 77/80 status of the nonshared patient-donor allotypes | |
| Matched | 955 (48%) |
| Mismatched | 996 (50%) |
| Unknown | 24 (1%) |
NA, not applicable.
Other malignancies included breast cancer, renal/kidney carcinoma, hepatoblastoma. Nonmalignancies included severe aplastic anemia, Shwachman-Diamond anemia, Diamond-Blackfan anemia, adrenoleukodystrophy, Wiskott Aldrich syndrome, hyper IgM syndrome, hemoglobinopathy, chronic granulomatous disease, familial erythro hemophagocytic lymphocytosis, paroxysmal nocturnal hemoglobinuria, metachromatic leukodystrophy, immunodysregulation polyendocrinopathy enteropathy X-linked like syndrome, Fanconi anemia, bone marrow aplasia, idiopathic bone marrow failure, sickle cell disease, immune deficiency disorder, thalassemia, inherited abnormalities of erythrocyte differentiation or function, other immune system disorders, inherited abnormality of platelets, inherited disorder of metabolism, histiocytic disorders and other nonmalignancies.
Additional characteristics are provided in supplemental Table 2.
Disease status prior to transplant is categorized as early (first complete remission [CR] of acute myeloid leukemia [AML] or acute lymphoblastic leukemia [ALL]; first chronic phase [CP] of CML; refractory anemia [RA]; refractory anemia with ring sideroblasts of myelodysplastic syndrome [MDS]; non-Hodgkin lymphoma in first CR; chronic lymphoid leukemia in CR); intermediate (second or higher CR of AML or ALL; second or higher CP or accelerated phase of CML; Hodgkin lymphoma in third CR); late or advanced (primary induction failure or first or higher relapse of AML or ALL; blast phase of CML; MDS RA with excess blasts or excess blasts in transformation; non-Hodgkin lymphoma in relapse; Hodgkin lymphoma in third relapse; myeloma; unnamed MDS or unknown).
Four individuals each encoded novel HLA-C sequences that have not yet been characterized using serological reagents.
HLA-A, -C, -B, -DRB1, and -DQB1 allotypes were typed at high resolution for 1959 pairs and medium resolution for 16 pairs.26 Fresh blood and skin, liver, and gastrointestinal biopsy specimens are not available for large-scale historic transplant recipients and unrelated donors for direct HLA-C expression analysis in peripheral blood and GVHD-affected organ sites. However, the legitimacy of the predictive algorithm for MFI values for each serologic-equivalent HLA-C*01-18 allotype has been described and is concordant between healthy cells and cells infected with HIV in vitro.22 Not only are expression levels reproducible, but predicted expression levels based on HLA-C allotype correlate strongly and significantly with observed expression values, the level of expression of a given HLA-C allotype is consistent across ethnic groups,22 and measurement of HLA-C transcript levels across allotypes replicates the same hierarchy of HLA-C cell surface expression levels.27 Because the MFI is a proxy for HLA-C expression levels and fresh cells are not available on historic transplant pairs, HLA-C expression levels were imputed using the MFI for each patient and donor HLA-C allotype according to previously published quantitative measurements of MFIs.22
Each pair was matched for 1 HLA-C (shared allotype) and mismatched for the second HLA-C (nonshared allotype). Each nonshared HLA-C mismatch was defined as an allele or antigen mismatch.3 Protein sequences of nonshared allotypes may have identical amino acids at certain hypervariable residues but different (nonidentical) amino acids at other residues.28 Any nonidentity at residues 116, 77, and 80 was determined by alignment to reference HLA-C sequences.28
Consent from patients and donors was obtained in accordance with the Declaration of Helsinki. The work was approved by the National Institutes of Health Office of Human Subjects Research Protections and the Institutional Review Board, Fred Hutchinson Cancer Research Center. HLA and clinical data were contributed by participants of the International Histocompatibility Working Group in Hematopoietic Cell Transplantation.26
Biostatistical analysis
We hypothesized that the level of expression of the patient’s mismatched allotype, the donor’s mismatched allotype, and/or the shared allotype influences the risks of grades III to IV acute GVHD, relapse, nonrelapse mortality (death without a preceding recurrence of the underlying disease), and overall mortality (henceforth mortality). Because previous studies demonstrated a correlation between the sum of the MFIs of the 2 HLA-C allotypes and HIV outcomes,22 we examined the sum of the MFIs of patients’ and donors’ HLA-C allotypes. Expression level was modeled as a continuous linear variable in Cox (for relapse, nonrelapse mortality, and mortality) and logistic regression models (for acute GVHD). Under this assumption, hazard ratios (HRs) and odds ratios (ORs) from these regression models are presented in terms of the increase in risk of failure associated with an increase in expression level of 100 fluorescence intensity units. All models were adjusted for age, source of stem cells, disease severity, T-cell depletion, and year of transplant (Table 1). Statistical interactions between expression level and mismatching defined according to 3 different models (allele vs antigen, residue 116, and residues 77/80) were assessed by including appropriate terms into regression models, and interactions between expression levels were assessed in the same way. Expression levels were dichotomized as low (C*07 and C*03) or high (C*01 and C*14) to demonstrate the interaction between expression level and mismatch model when appropriate. Mean expression levels were compared between groups for each mismatch model with the 2-sample t test. No adjustments for multiple comparisons were made, and P values between .01 and .05 should be considered suggestive, as opposed to conclusive, evidence of an association.
Results
The clinical characteristics of the patients and donors in the study population were consistent with the worldwide experience in unrelated donor hematopoietic cell transplantation (Table 1). Increasing expression level of the patient’s nonshared HLA-C was significantly associated with increased risks of acute GVHD, nonrelapse mortality, and mortality but not disease relapse (P = .003, .005, .009, and .76, respectively; Figure 1; Table 2). There was no suggestion that these effects differed across the various years of transplant included in the study for any of these end points (interaction tests between expression level and year of transplant, P = .56, .69, .42, and .55, respectively).
The level of expression of the patient’s mismatched HLA-C allotype associates with transplant outcome. (A) ORs of grades III to IV acute GVHD and (B) HRs of nonrelapse mortality for each mismatched HLA-C allotype in the patient is shown relative to C*07. The expression of the patient’s mismatched allotype was defined by its MFI as determined previously.22 The size of each circle is proportional to the number of patients with the indicated allotype. The least-squares line shown is weighted by the number of observations at each expression level.
The level of expression of the patient’s mismatched HLA-C allotype associates with transplant outcome. (A) ORs of grades III to IV acute GVHD and (B) HRs of nonrelapse mortality for each mismatched HLA-C allotype in the patient is shown relative to C*07. The expression of the patient’s mismatched allotype was defined by its MFI as determined previously.22 The size of each circle is proportional to the number of patients with the indicated allotype. The least-squares line shown is weighted by the number of observations at each expression level.
Level of HLA-C expression influences acute GVHD, nonrelapse mortality, and overall mortality, but not relapse
| Nonshared HLA-C allotype* . | Acute GVHD† (N = 453/1861) . | Nonrelapse mortality† (N = 709/1727) . | Overall mortality† (N = 1246/1975) . | Relapse† (N = 501/1727) . | ||||
|---|---|---|---|---|---|---|---|---|
| OR (95% CI) . | P . | HR (95% CI) . | P . | HR (95% CI) . | P . | HR (95% CI) . | P . | |
| Patient’s mismatch | 1.34 (1.10-1.62) | .003 | 1.22 (1.06-1.39) | .005 | 1.15 (1.03-1.27) | .009 | 1.03 (0.86-1.22) | .76 |
| Donor’s mismatch | 1.07 (0.88-1.30) | .49 | 1.15 (1.01-1.31) | .04 | 1.14 (1.03-1.26) | .01 | 0.97 (0.82-1.16) | .74 |
| Sum of mismatched allotypes | 1.16 (1.03-1.32) | .02 | 1.15 (1.06-1.25) | .002 | 1.12 (1.05-1.19) | .001 | 1.00 (0.89-1.12) | .96 |
| Nonshared HLA-C allotype* . | Acute GVHD† (N = 453/1861) . | Nonrelapse mortality† (N = 709/1727) . | Overall mortality† (N = 1246/1975) . | Relapse† (N = 501/1727) . | ||||
|---|---|---|---|---|---|---|---|---|
| OR (95% CI) . | P . | HR (95% CI) . | P . | HR (95% CI) . | P . | HR (95% CI) . | P . | |
| Patient’s mismatch | 1.34 (1.10-1.62) | .003 | 1.22 (1.06-1.39) | .005 | 1.15 (1.03-1.27) | .009 | 1.03 (0.86-1.22) | .76 |
| Donor’s mismatch | 1.07 (0.88-1.30) | .49 | 1.15 (1.01-1.31) | .04 | 1.14 (1.03-1.26) | .01 | 0.97 (0.82-1.16) | .74 |
| Sum of mismatched allotypes | 1.16 (1.03-1.32) | .02 | 1.15 (1.06-1.25) | .002 | 1.12 (1.05-1.19) | .001 | 1.00 (0.89-1.12) | .96 |
The level of expression of the patient’s mismatch, the donor’s mismatch, and the sum of these mismatched allotypes were each modeled as a continuous linear variable. ORs and HRs are presented as an increase in risk of failure associated with each increase in expression of 100 fluorescence intensity units.
For the shared (matched) allotype, the OR for acute GVHD was 0.90 (95% CI, 0.71-1.13; P = .36), the HR for nonrelapse mortality was 0.85 (95% CI, 0.72-1.00; P = .05), the HR for overall mortality was 0.94 (95% CI, 0.84-1.07; P = .34), and the HR for relapse was 1.10 (95% CI, 0.90-1.34; P = .34).
Numbers denote the number of patients with failure for each of the 4 end points of all patients with clinical data for the given end point.
Increasing expression level of the donor’s nonshared HLA-C was associated with increased nonrelapse mortality (P = .04) and mortality (P = .01), although with borderline significance. The sum of the MFIs of patients’ and donors’ nonshared allotypes was significantly associated with nonrelapse mortality (P = .002) and mortality (P = .001); there was no statistically significant association between donors’ expression level and acute GVHD or relapse (Table 2). The strong effect of patients’ HLA-C expression level on GVHD risk suggests enhanced graft-versus-host recognition of highly expressed patient allotypes by the donor graft.
Three mismatch models have been previously proposed to explain why some HLA-C mismatches are better tolerated than others.3-7,9-12,15-20 Because the expression level of the patient’s nonshared allotype correlated significantly with transplant outcome, we evaluated whether expression levels play a role in each of these mismatch models.
Model 1: allele and antigen mismatches
HLA-C allele mismatches may be better tolerated than antigen mismatches.3-7 We found striking differences in the mean HLA-C MFIs of the patients who had an allele mismatch relative to their donors (123.2 on average) compared with those who had an antigen mismatch relative to their donors (176.7 on average; P < .0001; Table 3), and the same was true for donors’ mismatched HLA-C allotypes (data not shown). The lower mean MFI of allele mismatches resulted from the overwhelming representation of common low-expression C*07 and C*03 mismatches (C*07:01/07:02; C*03:03/03:04), whereas antigen mismatches were more uniformly distributed across all expression levels. Thus, allele mismatches differ from antigen mismatches in the degree of sequence similarity3,5 and in expression levels, and these features may contribute to the historically lower risks of allele compared with antigen mismatches. These results are consistent with recent studies demonstrating that the HLA-C*03:03/03:04 mismatch is a well-tolerated high-frequency mismatch24,25 and provide a potential mechanism (ie, low expression) for why this particular mismatch is permissive.
Distribution of HLA-C allotypes according to 3 models of HLA-C mismatching
| Patient’s nonshared allotype* . | MFI . | HLA-C mismatch model for the nonshared patient allotype N (%) . | |||||
|---|---|---|---|---|---|---|---|
| Allele vs antigen† (N = 1971) . | Residue 116‡ (N = 1955) . | Residues 77/80§ (N = 1951) . | |||||
| Allele (N = 389) . | Antigen (N = 1582) . | Matched (N = 847) . | Mismatched(N = 1108) . | Matched (N = 955) . | Mismatched (N = 996) . | ||
| C*07 | 111 | 104 (27)¶ | 288 (18) | 242 (29) | 147 (13) | 210 (22) | 179 (18) |
| C*03 | 114 | 238 (61)|| | 187 (12) | 268 (32) | 155 (14) | 321 (34) | 100 (10) |
| C*17 | 115 | 0 | 3 (<1) | 1 (<1) | 2 (<1) | 1 (<1) | 2 (<1) |
| C*05 | 154 | 5 (1) | 147 (9) | 50 (6) | 100 (9) | 43 (5) | 107 (11) |
| C*02 | 164 | 1 (<1) | 146 (9) | 31 (4) | 114 (10) | 43 (5) | 102 (10) |
| C*08 | 176 | 5 (1) | 28 (2) | 10 (1) | 22 (2) | 15 (2) | 17 (2) |
| C*16 | 180 | 10 (3) | 91 (6) | 43 (5) | 57 (5) | 29 (3) | 71 (7) |
| C*12 | 193 | 4 (1) | 118 (7) | 84 (10) | 36 (3) | 86 (9) | 34 (3) |
| C*04 | 200 | 5 (1) | 161 (10) | 10 (1) | 155 (14) | 39 (4) | 126 (13) |
| C*15 | 223 | 11 (3) | 111 (7) | 4 (<1) | 118 (11) | 51 (5) | 71 (7) |
| C*06 | 225 | 5 (<1) | 63 (4) | 36 (4) | 32 (3) | 25 (3) | 43 (4) |
| C*18 | 239 | 0 | 2 (<1) | 1 (<1) | 1 (<1) | 1 (<1) | 1 (<1) |
| C*01 | 254 | 0 | 152 (10) | 40 (5) | 112 (10) | 68 (7) | 84 (8) |
| C*14 | 294 | 1 (<1) | 85 (5) | 27 (3) | 56 (5) | 23 (2) | 59 (6) |
| Mean MFI of the patient’s mismatched HLA-C allotype | 123.2 | 176.7 | 148.6 | 179.8 | 154.2 | 177.4 | |
| P < .0001 | P < .0001 | P < .0001 | |||||
| Patient’s nonshared allotype* . | MFI . | HLA-C mismatch model for the nonshared patient allotype N (%) . | |||||
|---|---|---|---|---|---|---|---|
| Allele vs antigen† (N = 1971) . | Residue 116‡ (N = 1955) . | Residues 77/80§ (N = 1951) . | |||||
| Allele (N = 389) . | Antigen (N = 1582) . | Matched (N = 847) . | Mismatched(N = 1108) . | Matched (N = 955) . | Mismatched (N = 996) . | ||
| C*07 | 111 | 104 (27)¶ | 288 (18) | 242 (29) | 147 (13) | 210 (22) | 179 (18) |
| C*03 | 114 | 238 (61)|| | 187 (12) | 268 (32) | 155 (14) | 321 (34) | 100 (10) |
| C*17 | 115 | 0 | 3 (<1) | 1 (<1) | 2 (<1) | 1 (<1) | 2 (<1) |
| C*05 | 154 | 5 (1) | 147 (9) | 50 (6) | 100 (9) | 43 (5) | 107 (11) |
| C*02 | 164 | 1 (<1) | 146 (9) | 31 (4) | 114 (10) | 43 (5) | 102 (10) |
| C*08 | 176 | 5 (1) | 28 (2) | 10 (1) | 22 (2) | 15 (2) | 17 (2) |
| C*16 | 180 | 10 (3) | 91 (6) | 43 (5) | 57 (5) | 29 (3) | 71 (7) |
| C*12 | 193 | 4 (1) | 118 (7) | 84 (10) | 36 (3) | 86 (9) | 34 (3) |
| C*04 | 200 | 5 (1) | 161 (10) | 10 (1) | 155 (14) | 39 (4) | 126 (13) |
| C*15 | 223 | 11 (3) | 111 (7) | 4 (<1) | 118 (11) | 51 (5) | 71 (7) |
| C*06 | 225 | 5 (<1) | 63 (4) | 36 (4) | 32 (3) | 25 (3) | 43 (4) |
| C*18 | 239 | 0 | 2 (<1) | 1 (<1) | 1 (<1) | 1 (<1) | 1 (<1) |
| C*01 | 254 | 0 | 152 (10) | 40 (5) | 112 (10) | 68 (7) | 84 (8) |
| C*14 | 294 | 1 (<1) | 85 (5) | 27 (3) | 56 (5) | 23 (2) | 59 (6) |
| Mean MFI of the patient’s mismatched HLA-C allotype | 123.2 | 176.7 | 148.6 | 179.8 | 154.2 | 177.4 | |
| P < .0001 | P < .0001 | P < .0001 | |||||
Patient allele mismatches as a group had significantly lower mean MFIs than patient antigen mismatches. The mean MFIs of the patients’ nonshared HLA-C allotypes were significantly different between allele and antigen mismatches, between residue 116 matches and residue 116 mismatches, and between residue 77/80 matches and residue 77/80 mismatches. Similar results were observed for donors’ nonshared allotypes (123.2 and 176.5, P < .0001 for allele and antigen mismatches; 147.0 and 179.9, P < .0001 for residue 116 matches and residue 116 mismatches; 150.5 and 180.2, P < .0001 for residue 77/80 matches and residue 77/80 mismatches). The mean MFIs of the shared matched allotypes, however, did not differ from one another (150.0 and 155.0, P = .06; 155.2 and 153.3, P = .44; 152.5 and 155.6, P = .15, respectively).
Patients’ mismatched (nonshared) allotypes are listed in order from lowest (C*07) to highest (C*14) MFI.22
HLA allele and antigens were defined according to WHO HLA Nomenclature.29 Four individuals each encoded novel HLA-C sequences that have not yet been characterized using serological reagents; these individuals were not included in the allele/antigen mismatch analysis.
Patients’ and donors’ nonshared HLA-C allotypes can be either matched or mismatched at residue 116. A total of 20 transplants were not included in the residue 116 analysis because they lacked sequence information at this position.
Patient’s and donors’ nonshared HLA-C allotypes can be either matched or mismatched at residues 77/80 that define the KIR C1 and C2 ligand groups. A total of 24 transplants were not included in the residue 77/80 analysis because they lacked sequencing information for residue 77 and/or 80 or did not have 77S-80N (C1) or 77N-80K (C2).
The most common patient-donor mismatch was C*07:01 vs C*07:02 or C*07:02 vs C*07:01; N = 79/104 (76%).
The most common patient-donor mismatch was C*03:03 vs C*03:04 or C*03:04 vs C*03:03; N = 216/238 (91%).
The difference in GVHD between antigen and allele mismatches in our study was not as large as previously reported6,7 (OR, 1.27; 95% confidence interval [CI], 0.96-1.68; P = .09). The virtual absence of higher expression allele mismatches precluded comparison of the effects of expression levels on outcome in allele- vs antigen-mismatched patients. However, because the full range of HLA-C allotypes was represented among patients mismatched for an HLA-C antigen (Table 3), the importance of expression on outcome can be defined in antigen-mismatched transplants, thereby removing the allele/antigen effect. Among patients mismatched for 1 HLA-C antigen, increasing expression level of the patient’s mismatched HLA-C was associated with increased risk of acute GVHD (OR, 1.36; 95% CI, 1.09-1.69; P = .006).
Moreover, among patients mismatched for the lowest expressing allotypes (C*07, C*03), the risk of GVHD was similar between antigen and allele mismatches (OR, 1.07; 95% CI, 0.75-1.53; P = .70). These results suggest that antigen mismatches are tolerated if the patient’s mismatched antigen is expressed at low levels.
The risk of nonrelapse mortality was increased for antigen vs allele mismatches but was not statistically significant (HR, 1.16; 95% CI, 0.95-1.41; P = .13); the risk of mortality was also marginally increased (HR, 1.17; 95% CI, 1.01-1.35; P = .04). Among antigen mismatches, nonrelapse mortality was significantly increased (HR, 1.23; 95% CI, 1.06-1.44; P = .007), as was mortality, albeit with borderline significance (HR, 1.13; 95% CI, 1.01-1.27; P = .03) as the expression level of the patient’s mismatched HLA-C increased. Among the lowest expression mismatches (C*07, C*03), the risk of nonrelapse mortality was similar among antigen and allele mismatches (HR, 1.06; 95% CI, 0.83-1.35; P = .66) as was the risk of mortality (HR, 1.11; 95% CI, 0.93-1.33; P = .26).
These data suggest that risks may be defined more by the expression level of the patient’s mismatched allotype than by the allele or antigen designation. Low-expression antigen mismatches appear to be as readily tolerated as allele mismatches.
Model 2: residue 116 mismatches
Residue 116 plays a key role in determining the peptide repertoire of class I allotypes,8,13,14 and patient-donor mismatching at this position may influence transplant outcome.9-12 The mean MFIs of patients’ (and donors’) nonshared allotypes were statistically significantly lower among residue-matched compared with residue-mismatched pairs (Table 3); the mean MFIs for the shared allotype were similar. These results might explain the observation of lower risks after residue-matched compared with residue-mismatched transplantation.9-12
Residue-mismatched patients had an increased risk of acute GVHD compared with residue-matched patients, but the difference was not statistically significant (OR, 1.14; 95% CI, 0.91-1.41; P = .26). There was no evidence to suggest that the association between expression level and GVHD was different among residue-mismatched and residue-matched patients (interaction, P = .55); therefore, separate analyses of expression levels and GVHD risk were not conducted for these groups.
The risk of overall mortality was slightly higher in residue-mismatched patients (HR, 1.12; 95% CI, 1.00-1.26; P = .05), but an interaction test between expression levels and mismatching was not statistically significant (P = .21).
The risk of nonrelapse mortality was higher among residue-mismatched patients than among residue-matched patients with borderline significance (HR, 1.21; 95% CI, 1.04-1.40; P = .02), but the impact of residue mismatching on nonrelapse mortality depended on the expression level (interaction, P = .05). Specifically, nonrelapse mortality increased as expression levels increased among residue-mismatched patients (HR, 1.31; 95% CI, 1.09-1.58; P = .004), but not among residue-matched patients (HR, 0.98; 95% CI, 0.78-1.23; P = .88).
One specific demonstration of this interaction is seen when patients whose nonshared allotype falls at the lower end of the MFI scale (C*07 or C*03, or low expression) are compared with those whose nonshared allotype falls at the higher end of the MFI scale (C*01 or C*14, or high expression). The negative impact of high expression (relative to low expression) on nonrelapse mortality was seen among residue-mismatched patients (HR, 1.64) but not residue-matched patients (HR, 0.85). Similarly, the negative impact of residue mismatching (relative to residue matching) on nonrelapse mortality was observed among patients with high-expression mismatches (HR, 1.97) but not among patients with low-expression mismatches (HR, 1.02).
These results were virtually identical for antigen-mismatched patients (data not shown), and they suggest that the increased risk of nonrelapse mortality associated with high-expression mismatches is confined to patients who are mismatched at residue 116 and vice versa.
Model 3: residue 77/80 mismatches
The mean MFIs of nonshared allotypes were significantly lower among residue 77/80-matched compared with residue 77/80-mismatched patients (Table 3) and might explain lower risks with residue 77/80 matching in some studies.15-20
There was no association between residue 77/80 mismatching and acute GVHD (OR, 1.02; 95% CI, 0.82-1.27; P = .83). The magnitude of the association between the expression level of the patient’s mismatched HLA-C and GVHD was larger among residue 77/80-mismatched patients than among residue 77/80-matched patients, but this difference was not statistically significant (interaction, P = .11), and therefore, separate analyses of residue 77/80 matching are not presented.
The risk of overall mortality was suggestively higher in residue 77/80-mismatched patients (HR, 1.13; 95% CI, 1.01-1.26; P = .04), but an interaction test between expression and mismatching was not statistically significant (P = .20).
There was no statistically significant association between residue 77/80 mismatching and the risk of nonrelapse mortality (HR, 1.09; 95% CI, 0.94-1.27; P = .24); however, the association between residue 77/80 mismatching and nonrelapse mortality appeared to depend on the expression level of the patient’s nonshared HLA-C (interaction, P = .02). In particular, nonrelapse mortality increased as expression levels increased among residue 77/80-mismatched patients (HR, 1.38; 95% CI, 1.14-1.67; P = .0009), but not among residue 77/80-matched patients (HR, 1.01; 95% CI, 0.82-1.24; P = .91).
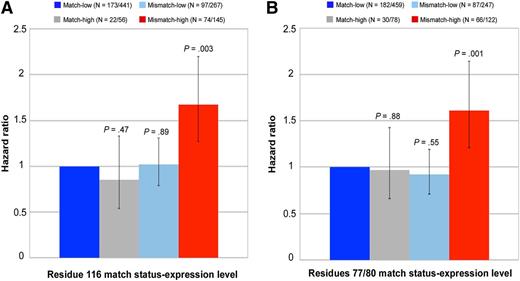
Similar to residue 116, evaluation of patient mismatches with extreme expression levels provides a specific demonstration of the interaction between expression level and residue 77/80 match status (Figure 2). The negative effect of high expression (relative to low MFI) on nonrelapse mortality was evident among residue 77/80-mismatched patients (HR, 1.74) but not residue 77/80-matched patients (HR, 0.97); similarly, the negative impact of residue 77/80 mismatching (relative to residue 77/80 matching) was observed among patients with high-expression (HR, 1.66) but not low-expression mismatches (HR, 0.92). When restricted to patients with antigen mismatches, the results were qualitatively the same (data not shown). These data suggest that MFIs provide information on the permissiveness of residue 77/80 mismatches. Whereas low- or high-expression residue 77/80 matches and low-expression residue 77/80 mismatches are well tolerated (permissible), high-expression residue 77/80 mismatches are not permissible.
Demonstration of interaction between expression level of patients’ nonshared HLA-C and residue 116 and residue 77/80 match status for nonrelapse mortality. Patients’ low-expression mismatched allotypes were defined as HLA-C*07 and C*03; high-expression mismatched allotypes were defined as HLA-C*01 and C*14. Each 95% CI is denoted by black bars. Numbers in parentheses indicate the patients who died of nonrelapse causes of the total number of patients in each of the 4 groups as defined by residue 116 match status (A), residue 77/80 match status (B), and level of expression of the patients’ mismatched HLA-C.
Demonstration of interaction between expression level of patients’ nonshared HLA-C and residue 116 and residue 77/80 match status for nonrelapse mortality. Patients’ low-expression mismatched allotypes were defined as HLA-C*07 and C*03; high-expression mismatched allotypes were defined as HLA-C*01 and C*14. Each 95% CI is denoted by black bars. Numbers in parentheses indicate the patients who died of nonrelapse causes of the total number of patients in each of the 4 groups as defined by residue 116 match status (A), residue 77/80 match status (B), and level of expression of the patients’ mismatched HLA-C.
The impact of expression levels on grades III to IV acute GVHD was similar when other variables were taken into account such as source of stem cells, use of T-cell depletion, year of transplant, and severity of disease (ie, no statistically significant interactions were observed between expression level and any of these factors). Collectively, all 3 models of transplant alloreactivity demonstrate that the expression level of the patient’s nonshared HLA-C allotype is associated with transplant outcome.
Discussion
An unmet need in transplantation is a functional measure of HLA-C mismatching that can be shown to be associated with clinical outcome. The availability of such a tool would provide options for the use of selected HLA-C-mismatched unrelated donors without increasing life-threatening acute GVHD and mortality. The recent discovery that HLA-C expression levels have direct consequences on HIV-AIDS progression and susceptibility to Crohn disease22 provides a framework for studying HLA-C in transplantation. To test our hypotheses, we leveraged an expansive international collaboration among immunogenetic laboratories, transplant centers, and transplant/donor registries to identify patients who have only 1 HLA-C mismatch with their donor. This unique genetic relationship between the transplant patient and donor permitted the effect of HLA-C expression levels to be examined for single HLA-C molecules.
The collection of archived DNA samples from patients and donors is a unique and precious international scientific resource. As is true for virtually all large retrospective disease cohorts, no viable cells are available for in vitro HLA cell surface measurements of expression. Notably, such measurements require fresh cells, as freezing causes a significant artifactual decrease in HLA expression. Given the shortfalls of direct in vitro measurement of HLA-C expression in historic transplant study subjects, the use of inferred expression data is needed. As previously demonstrated, HLA-C expression levels, as defined by MFI, correlate with the HLA-C allotype.22 Use of an HLA-C-specific antibody that binds all HLA-C allotypes equally23 has shown that, in a panel of fresh cells from both African Americans and European Americans, HLA-C expression levels reproducibly correlate strongly and significantly with the HLA-C allotype, and the expression level of a given allotype is consistent across diverse populations.22 Furthermore, HLA-C transcript levels across allotypes replicate the same hierarchy of HLA-C expression levels.27 Given these results and the lack of the ability to directly measure HLA-C expression levels, we used MFI as a proxy for expression and examined the association of MFI with outcome.
The MFI is superior to any other genetic proxy for HLA-C expression when studying disease association22,23,27,30,31 and provides allotype-specific information for each transplant patient and donor. We found that the expression level of the patient’s mismatched HLA-C is a key determinant of transplant outcome, where mismatches involving alleles that have, on average, higher expression levels are more poorly tolerated than mismatches involving alleles that have, on average, lower expression levels. Moreover, the expression levels of patients’ mismatched allotypes distinguish permissible (lower MFI) from nonpermissible (higher MFI) antigen, residue 116, and residue 77/80 mismatches. Hence, HLA-C expression may introduce a new principle to the paradigm of alloreactivity in transplantation.
The graft-versus-leukemia effect describes a lower risk of relapse among patients with clinical GVHD compared with patients without GVHD.32 In the current analysis, we observed higher risks of acute GVHD, nonrelapse mortality, and mortality associated with increasing expression levels of the patient’s nonshared HLA-C, without a lowered risk of disease relapse. These results suggest that GVHD-independent relapse may involve the level of surface expression of HLA and may help to explain why some patients relapse despite developing clinical GVHD. The role of HLA expression in the graft-versus-leukemia effect merits further investigation in the future, when sufficiently large numbers of patients that encode the full range of HLA-C allotypes can be examined.
We surmise that mismatched allotypes expressed at lower levels are more likely to escape detection by the donor, as indicated by the decreased risk of poor outcomes among patients who are mismatched for low-expression allotypes. Our results suggest that avoidance of mismatching for higher expression allotypes in patients may help to lower overall risks after transplantation. When matched donors are not available, mismatching for the lower-expression allotype in the patient may lower risks of GVHD and mortality. Avoidance of HLA-C-mismatched donors altogether for patients with 2 highly expressed allotypes is advisable.
The residue mismatch model posits that donor T cells recognize differences in the HLA class I/peptide complex of patients.8,13,14 When HLA-C mismatches involve differences at residues 77/80, they may provoke recognition of donor KIR for patient’s HLA-C ligands that are not self.15 A given HLA-C allotype in the patient could generate in vivo T-cell, as well as NK, responses from the donor, but the balance of T and NK reactivity may depend on many factors including those that influence the maturation of NK cells in the donor.33-35 In HLA-C-mismatched unrelated donor transplantation, both T- and NK-mediated allorecognition may contribute to transplant-associated risks. Patient-donor variation at several key residues of class I molecules might be associated with transplant outcome, and of these, residue 116 has received particular attention because of its importance in the peptide-anchoring F pocket of the class I molecule and its high frequency of mismatching among transplant pairs.9,10,12 Residues 77 and 80 reside with residue 116 in the F pocket and influence the size, shape, and charge of the peptide-binding groove and the carboxy-terminal peptide anchor.8,13,14 Hence, the same residues that define the cognate ligands of KIRs may also influence the nature of the HLA-C/peptide complex. As any residue 77/80 mismatch is also an allele or antigen mismatch, the specific contribution to nonrelapse mortality by T or NK pathways is almost certainly intertwined. The importance of patient-donor mismatching at other class I residues is also of interest. Many HLA-C mismatch combinations involve concurrent mismatching at multiple residues, but few examples exist in the clinical population where allotypes differ at only 1 of these positions, which is essential for appropriate comparison of residue-specific risks. In addition to residue-specific effects, the role of expression in influencing the immunogenicity of a given HLA allotype in the context of its repertoire of minor histocompatibility antigens is an important question. Such studies may be feasible in the future if and when a sufficiently larger transplant experience is available.
This study provides new insight into the strength of the immune response of HLA-C in transplantation and a platform for exploring the mechanistic basis of T-cell and NK recognition of HLA-C allotypes. Application of the findings in the current study can be envisioned for future patients who do not have HLA-matched donors as an option. The effects of differential allotype expression levels at other HLA loci may further delineate and broaden the pool of acceptable donors for patients, and given the results presented herein, characterizing such effects is warranted.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
E.W.P., T.A.G., M.M., S.R.S., M.D.H., and M.M.H. are supported by National Institute of Allergy and Infectious Diseases (NIAID) grant AI069197; E.W.P., T.A.G., and M.M. are supported by National Cancer Institute (NCI) grant CA18029; M.M.H. is supported by NCI grant CA76518 and Health Resources and Services Administration grant HHSH234200637015C; S.R.S., M.D.H., and M.M.H. are supported by NCI grant U24-CA76518, the National Heart, Lung and Blood Institute (NHLBI), NIAID, NHLBI and NCI grant 5U01HL069294, and Office of Naval Research grants N00014-12-1-0142 and N00014-13-1-0039; A.P.B. was supported by The Associazione Italiana Ricerca sul Cancro (Milano, Italy); P.J. was supported by Project ED2.1.00/03.0076 from the European Regional Development Fund, T.A. ČR TA01010342, and GACR NT/12454-5, Czech Republic; O.R. was supported by The Swedish Cancer Society, The Swedish Research Council, The Children’s Cancer Foundation, The Cancer Society in Stockholm, and Karolinska Institutet (Stockholm, Sweden); J.-M.T. was supported by Swiss National Science Foundation grant 310030-146306/1; and M.C. was supported by Frederick National Laboratory for Cancer Research contract HHSN261200800001E and the Intramural Research Program of the National Institutes of Health, Frederick National Laboratory, Center for Cancer Research.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government. The views expressed in this article do not reflect the official policy or position of the National Institutes of Health, the Department of the Navy, the Department of Defense, or any other agency of the US Government. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Authorship
Contribution: E.W.P. conceived and designed the experiments; E.W.P. and M.C. performed the experiments; T.A.G. analyzed the data; E.W.P., A.P.B., A.C., E.D.T., G.E., T.E., G.F.F., T.G., M.D.H., M.M.H., K.H., P.J., A.M., M.O., O.R., M.L.S., S.R.S., J.-M.T., A.V., and C.S.W. contributed reagents/materials/analysis tools; M.M. managed data; E.W.P. wrote the first draft of the manuscript; E.W.P., T.A.G., and M.C. contributed to the writing of the manuscript; and M.C., C.O., and R.A. contributed MFI values and intellectual content.
A complete list of the members of the International Histocompatibility Working Group in Hematopoietic Cell Transplantation investigators is provided in supplemental Table 1, available on the Blood Web site.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Effie W. Petersdorf, Fred Hutchinson Cancer Research Center, 1100 Fairview Ave North, D4-115, PO Box 19024, Seattle, WA 98109; e-mail: epetersd@fhcrc.org.