Abstract
Acute myeloid leukemia (AML) is one of the most common and fatal forms of hematopoietic malignancies with diverse chromosomal and molecular abnormalities. The majority of AML patients do not survive more than 5 years. Advanced genomic studies reveal that both genetic and epigenetic abnormalities frequently occur in de novo AML. However, it remains a challenge to understand the complicated genetic/epigenetic regulatory networks and identify the functionally important nodes in these networks. There is an urgent need to develop effective therapeutic strategies based on these new insights.
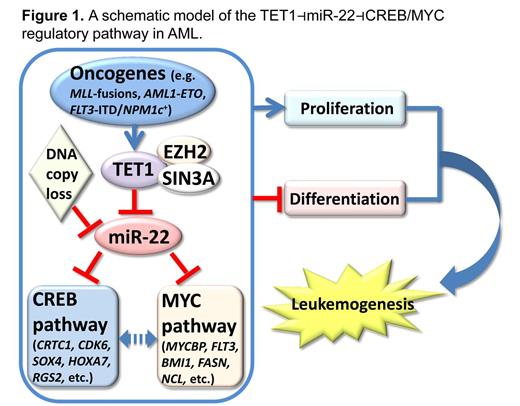
The ten-eleven translocation (Tet) proteins are important epigenetic regulators, which can convert 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) and lead to DNA demethylation. Among the three TET family members (TET1/2/3), TET2 was identified as a tumor suppressor in myeloid malignancies. Our lab recently reported that TET1 is highly expressed in MLL/KMT2A (Mixed Lineage Leukemia)-rearranged AML, a subtype of AML with poor prognosis. It is a direct target activated by MLL-fusions, and functions as an essential oncogene (Huang et al., PNAS, 2013). However, the function and regulatory pathway(s) of TET1 in AML remain poorly understood.
MicroRNAs (miRNAs) are a class of small, non-coding RNAs that play important roles in posttranscriptional gene regulation. Dysregulation of miRNAs is frequently observed in AML. Results of our profiling assays show that miR-22 is widely down-regulated in all major subtypes of de novo AML (Jiang et al., Cancer Cell, 2012), implying a tumor suppressor function. However, an oncogenic role for miR-22 was recently reported in myelodysplastic syndromes (MDS) and breast cancer, in which TET2 was repressed by miR-22 as its direct target gene.
Here we show that, amongst a group of miRNAs (e.g. miR-495 and miR-150, etc.) whose expression levels are repressed in AML, miR-22 exhibits the most potent and consistent inhibition on MLL-AF9-induced transformation of mouse bone marrow (BM) progenitor cells. Moreover, forced expression of miR-22 dramatically inhibits cell transformation and leukemogenesis induced by multiple fusion genes, such as MLL-fusions and RUNX1/AML1-ETO9a. Furthermore, the maintenance of various subtypes of AML (e.g., those induced by MLL-fusion, AML1-ETO9a or FLT3-ITD/NPM1c+) is also dependent on the repression of miR-22. Thus, our data demonstrate a potent tumor-suppressor role of miR-22 in AML.
Surprisingly, our analysis of three (in-house and outside) large-scale AML datasets revealed that TET2 (and likely also TET3) expression levels exhibited a significant positive correlation, whereas only TET1 exhibited a significant negative correlation (r<-0.32; p<0.001), with miR-22 expression. Our subsequent ChIP/qPCR studies suggest an epigenetic repression on miR-22 transcription mediated by TET1 and its repressive cofactors such as SIN3A and EZH2, through their direct binding to the miR-22 promoter region and subsequent modifications of histone markers such as H3K27Me3 which, in turn, inhibit RNA polymerase II recruitment and, thereby, miR-22 transcription. Besides the TET1-mediated epigenetic repression, the miR-22 locus (within 17p13.3) is also affected by DNA copy loss in 8-20% of AML cases, further highlighting its tumor-suppressor role in AML.
Further, through a series of data analyses followed by experimental validations and functional studies, we show that a set of critical oncogenes, including CRTC1, FLT3 and MYCBP, are functionally important direct target genes of miR-22 in AML and thus, miR-22 negatively regulates the CREB and MYC signaling pathways. Our proof-of-concept study shows that miR-22 RNA oligos formulated with dendritic nanoparticles significantly inhibit leukemia progression and extend the overall median survival of MLL-AF9-induced leukemic mice from 29 days to 54 days (n=10 per group, p<0.001, log-rank test) in mouse BM transplantation assays, indicating the therapeutic potential of miR-22 in treating AML.
Taken together, our results demonstrate a potent tumor-suppressor role of miR-22 in AML, and suggest the potential clinical application of miR-22-nanoparticles in treating AML. We also identified a TET1⊣miR-22⊣CREB/MYC regulatory pathway, which is critical in AML pathogenesis (see Fig. 1). Our findings also highlight potential distinct genetic/epigenetic mechanisms underlying de novo AML and MDS.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal