Abstract
Purpose: Chronic myelomonocytic leukemia (CMML) is a clinically heterogeneous myelodysplastic/myeloproliferative neoplasm with short overall survival. Emerging data based on sequencing of candidate genes with a known role in myeloid leukemia have identified recurrent CMML mutations, some of which have been associated with poor prognosis. A comprehensive evaluation of the mutational landscape of CMML and its prognostic significance is lacking.
Patients and Methods. We comprehensively characterized the mutational landscape of CMML by a 2-step design. We initially performed whole exome sequencing (WES) of paired leukemia and germline DNA from 21 patients with a confirmed CMML diagnosis (discovery cohort) to identify genes with somatic mutations. From this discovery cohort, 215 genes showing potential mutations as well as 61 genes selected from the literature were examined by targeted resequencing in a second cohort of 69 clinically annotated CMML patients, using two independent platforms for orthogonal confirmation Blood or marrow samples from 22 young and 17 old controls were included as controls.
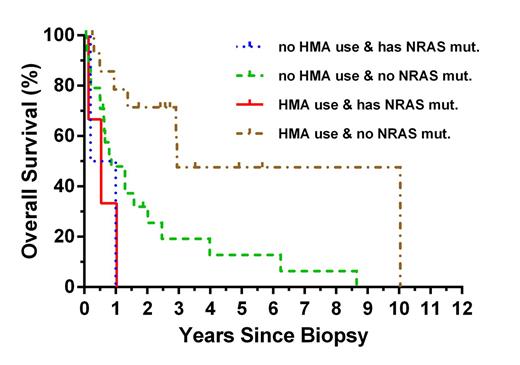
Results. We identified 22 genes with mutations in >3% of CMML patients, and 67/69 patients (97%) had one or more mutations in at least one of these genes. SRSF2, ASXL1 and TET2 were the most frequently mutated genes. Several novel CMML genes were identified, including FAT4 with a mutation prevalence of 10% and BCR, CBFA2T3, and TRPM1 with a prevalence of 3 – 9% (see Table). Total deleterious mutations per patient ranged from 0 – 11, with significant exclusion or association of various combinations of mutations in SETBP1, ASXL1, KRAS, TET2, and EZH2 observed. In univariate analysis hemoglobin < 9g/dL, white blood cell count > 15x109/L, no treatment with hypomethylating agents, mutations in ASXL1, EZH2, or NRAS and mutations in growth factor signaling genes were associated with shorter overall survival. In multivariate analysis, mutations in NRAS (P<0.05) and lack of therapy with hypomethylating agents (P<0.005) retained statistical significance (see Figure). In additional exploratory analyses mutated genes were grouped according to function. Patients with mutations in genes related to growth factor signaling had significantly shorter survival (P<0.005), while mutations in other functional groups were not predictive of outcome.
Conclusion. (i) The mutational landscape of CMML is complex and involves mutations in multiple genes, many affected with relatively low prevalence. (ii) Mutations in FAT4, a putative tumor suppressor and key regulator of the Hippo pathway, occur in approximately 10% of patients. (iii) Absence of therapy with hypomethylating agents and mutations in NRAS or the functional group of growth factor signaling genes are predictive of poor survival.
Count and prevalence of N=69 CMML patients having a deleterious mutation in one of the genes observed mutated at a prevalence ≥ 10%.
| Gene . | # CMML Patients with Mutation (%) . |
|---|---|
| SRSF2 | 34 (49%) |
| TET2 | 28 (41%) |
| ASXL1 | 25 (36%) |
| RUNX1 | 14 (20%) |
| SETBP1 | 11 (16%) |
| KRAS | 10 (14%) |
| EZH2 | 8 (12%) |
| FAT4 | 7 (10%) |
| CBL | 7 (10%) |
| NRAS | 7 (10%) |
| Gene . | # CMML Patients with Mutation (%) . |
|---|---|
| SRSF2 | 34 (49%) |
| TET2 | 28 (41%) |
| ASXL1 | 25 (36%) |
| RUNX1 | 14 (20%) |
| SETBP1 | 11 (16%) |
| KRAS | 10 (14%) |
| EZH2 | 8 (12%) |
| FAT4 | 7 (10%) |
| CBL | 7 (10%) |
| NRAS | 7 (10%) |
Overall survival of N=48 CMML patients by HMA use and presence of deleterious mutation in NRAS.
Overall survival of N=48 CMML patients by HMA use and presence of deleterious mutation in NRAS.
Mason:Agilent, Inc.: Research Funding. Deininger:Celgene: Research Funding; Agilent, Inc.: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal