Abstract
Background: Immunoglobulin (Ig) gene rearrangement is a hallmark of early B-cell development. Chronic lymphocytic leukemia (CLL) is typically considered a malignancy of mature B-cells and is thought to originate from the oncogenic transformation of a single pre- or post-germinal B-cell. Activation-induced deaminase (AID), an enzyme that induces somatic hypermutation (SHM) at the heavy and light chain Ig loci, has been shown to be active in CLL cells in vitro (Patten et al., Blood 2012). Previous studies suggest that multiple CLL-specific Ig clonotypes related by SHM may be present in patients (pts) with dominant CLL clones possessing somatically mutated or unmutated Ig loci (Logan et al., PNAS 2011; Campbell et al., PNAS 2008). To our knowledge, evolution of the dominant CLL-specific Ig clonotype over the course of treatment has not been demonstrated. Here we utilized the LymphoSIGHT™ method, a next-generation sequencing-based method for lymphocyte characterization and quantification, to quantify clonal evolution at the Ig heavy and kappa chain (IGH and IGK) loci in 63 pts with CLL.
Methods: Samples were collected at Stanford University and the Dana-Farber Cancer Institute. Peripheral blood mononuclear cells were isolated, and genomic DNA was extracted. Using unbiased universal primer sets, we amplified IGH and IGK variable, diversity, and joining gene segments. Amplified products were sequenced and analyzed using standardized algorithms for clonotype determination (Faham et al., Blood 2012). CLL-specific clonotypes were identified for each patient based on their high frequency (>5%) within the B-cell repertoire of a diagnostic (dx) sample. The highest frequency CLL clonotype identified in a dx sample is termed the “index clonotype”. Dx and post-treatment peripheral blood samples were assessed for evidence of evolved CLL clonotypes using LymphoSIGHT. A clonotype was considered “evolved” based on CDR3 sequence homology to the dx “index clonotype.”
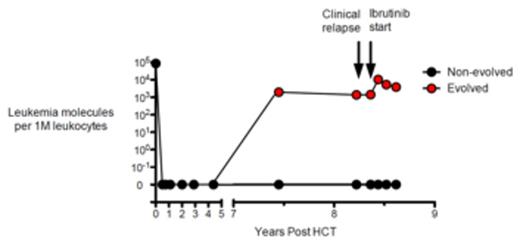
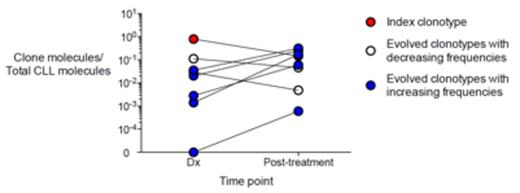
Results: CLL clonotypes were identified in dx samples from 63 pts (51 unmutated IGHV; 12 mutated), and we assessed post-treatment samples for the presence of CLL clonotype-associated oligoclonality. Two of 63 pts exhibited clonal evolution in post-treatment samples. One patient with unmutated CLL was MRD negative for over 7 years following allogeneic hematopoietic cell transplant (HCT), and subsequently became MRD positive with the evolved clonotype (differing by 1 nucleotide from the index clonotype) leading to clinical relapse 9 months after MRD positivity, while the original index clone remained undetectable. The patient was treated with ibrutinib upon clinical relapse and continues to have detectable MRD with the same evolved CLL clonotype (Fig 1A). In a second patient with mutated IGHV, we observed several evolved clonotypes in the dx sample. Multiple evolved clonotypes, including 5 that exhibited a significant increase in their frequency relative to the index clonotype, were present in the follow-up sample after treatment with fludarabine and rituximab (Fig 1B). These evolved clonotypes differed from the index clonotype by 1-4 nucleotides, but otherwise shared CDR3 identity, excluding independently arisen B cell clonotypes.
Conclusions: We observed evidence of clonal evolution at Ig loci in a small subset (3.2%) of pts with CLL undergoing treatment. The presence of evolution in pts with CLL indicates that either the SHM mechanism, including the AID enzyme, remains active after neoplastic transformation, or the evolved clonotypes arose through a mechanism distinct from SHM. These evolved CLL clonotypes may have a selective advantage, and may be useful as surrogate markers for other oncogenic mutations providing resistance to therapy. Additional cases are under investigation and updated results will be presented.
Figure 1. CLL clonal evolution during therapy. MRD levels of two related Ig clonotypes, expressed as leukemia molecules per million leukocytes in peripheral blood, are shown at multiple time points following allogeneic HCT (A). In another patient undergoing conventional treatment, the level of each individual evolved clonotype as a fraction of the total CLL molecules is plotted at dx and post treatment time points. The index clone, evolved clones with increasing levels post-treatment, and evolved clones with decreasing levels post-treatment are shown in red, blue, and white, respectively (B).
Moorhead:Sequenta, Inc.: Employment, Equity Ownership. Carlton:Sequenta, Inc.: Employment, Equity Ownership. Faham:Sequenta, Inc.: Employment, Equity Ownership, Membership on an entity's Board of Directors or advisory committees.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal