Abstract
Background:
Several retrospective series have characterized the genomic landscape in many lymphoma subtypes. However, as these studies used different sequencing methods, depth of coverage, and specimen source, the incidence of alterations in many lymphoma subtypes remain unknown. Even when common genomic alterations were identified within a lymphoma subtype, the incidence and spectrum of the alterations varied. In this study, we prospectively examined the incidence of genomic alterations across different lymphoma subtypes using the Foundation One Heme (FOH) assay.
Methods:
FOH testing was offered to patients in routine clinical practice. Formalin-fixed paraffin-embedded (FFPE) tissue, bone marrow aspirate or peripheral blood were examined using the FOH targeted sequencing assay. Hybridization capture from 405 cancer related genes and 31 genes commonly rearranged in cancer was applied to ≥ 50ng of DNA extracted from 49 tumor specimens and sequenced to high, uniform coverage. Genomic alterations, which include: base substitutions, small insertions and deletions (indels), rearrangements, and copy number alterations, were determined.
Results:
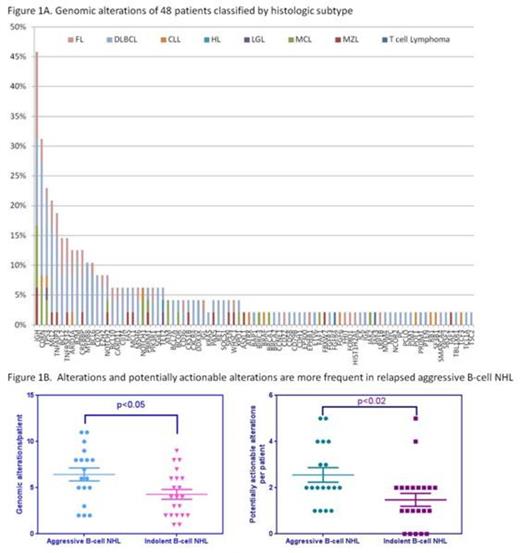
Specimens from 48 non-consecutive patients with lymphoma (5 new diagnoses, 42 relapsed) were prospectively examined. Represented subtypes include DLBCL (44%), FL (17%), CLL (10%), MCL (10%), MZL (8%), T cell lymphoma (4%), and Hodgkin lymphoma (2%). Median age of diagnosis was 48.1 years (range 29.5-80.6) with a male predominance (69%). The majority (75%) had advanced stage disease at diagnosis, with a median of 3 lines of therapy (range 1-12, n=44). Forty-nine specimens, (1 patient with 2 specimens) were collected from FFPE (70%), peripheral blood (22%) or bone marrow aspirate (8%). Reports resulted after a median 28 days (range 10-48 days) from the time specimens were shipped to Foundation Medicine.
Across 48 patients, 90 distinct alterations were detected with a median of 5 alterations/patient (range 0-11). Genomic alterations were more commonly observed in relapsed aggressive lymphoma compared to relapsed indolent lymphoma (median 6.4 vs 4 alterations/patient, p<0.05) or in newly diagnosed patients. The most common alteration was various IgH translocations (46%) and alterations in cyclin dependent kinases (CDK) genes (CDKN2A, CDKN2B and CDKN2C) (31%). Alterations of p53 (23%) and MLL2 (21%) were also frequently observed. Several rare alterations not previously described in primary lymphomas were identified, including alterations in ATRX, CDH1, MSH6, pyruvate carboxylase, and thrombospondin receptor (CD36). Alterations associated with approved agents or clinical trials were found in 89% of patients and were more frequent in relapsed aggressive B-cell NHL (median 2/patient) versus relapsed indolent B-cell NHL (median 1/patient, p<0.02). Commonly identified actionable alterations included CDK alterations in DLBCL, MCL and CLL, and alterations in the PI3K oncogenic pathway in DLBCL, FL and MZL.
Conclusion:
Application of a comprehensive next generation genomic sequencing assay provides an opportunity to both describe the spectrum and compare the incidence of genetic alterations across different lymphoma subtypes. Preliminary data suggest the vast majority of patients have one or more genomic alterations linked to approved agents or clinical trials. Data collection is ongoing. While the true incidence of each genomic alteration is not defined, the dataset provides the frequency of genomic alterations in a clinically relevant population. Moreover, these data will facilitate design of clinical trials by providing the opportunity to select patients based on shared genomic alterations rather than lymphoma subtype.
Clinical characteristics, N=48
| Age at diagnosis . | 57.1 (29.5-80.6) . |
|---|---|
| N (% of patients) | |
| Sex Male Female | 33 (68.7%) 16 (33.3%) |
| Subtype DLBCL | 21 (43.8%) |
| FL | 8 (16.7%) |
| CLL | 5 (10.4%) |
| MCL | 5 (10.4%) |
| MZL | 4 (8.3%) |
| T cell lymphoma | 2 (4.2%) |
| Hodgkin lymphoma | 1 (2.1%) |
| Other | 3 (6.3%) |
| Stage at Diagnosis I/II III/IV Not recorded | 10 (20.8%) 36 (75.0%) 2 (4.2%) |
| Biopsy sent At diagnosis At relapse | 6 (12.5%) 42 (87.5%) |
| Response to First line Therapy Near CR or CR PR SD PD Unknown | 22 (45.8%) 9 (18.8%) 5 (10.4%) 3 (8.3%) 8 (16.7%) |
| Genomic Alterations with clinical value Potentially Prognostic Potentially Actionable Alterations previously not well described | 37 (77.0%) 43 (89.6%) 7 (14.6%) |
| Age at diagnosis . | 57.1 (29.5-80.6) . |
|---|---|
| N (% of patients) | |
| Sex Male Female | 33 (68.7%) 16 (33.3%) |
| Subtype DLBCL | 21 (43.8%) |
| FL | 8 (16.7%) |
| CLL | 5 (10.4%) |
| MCL | 5 (10.4%) |
| MZL | 4 (8.3%) |
| T cell lymphoma | 2 (4.2%) |
| Hodgkin lymphoma | 1 (2.1%) |
| Other | 3 (6.3%) |
| Stage at Diagnosis I/II III/IV Not recorded | 10 (20.8%) 36 (75.0%) 2 (4.2%) |
| Biopsy sent At diagnosis At relapse | 6 (12.5%) 42 (87.5%) |
| Response to First line Therapy Near CR or CR PR SD PD Unknown | 22 (45.8%) 9 (18.8%) 5 (10.4%) 3 (8.3%) 8 (16.7%) |
| Genomic Alterations with clinical value Potentially Prognostic Potentially Actionable Alterations previously not well described | 37 (77.0%) 43 (89.6%) 7 (14.6%) |
Moskowitz:Seattle Genetics, Inc.: Consultancy, Research Funding; Genentech: Research Funding; Merck: Research Funding. Horwitz:Celgene: Consultancy, Research Funding; Millennium: Consultancy, Research Funding; Infinity: Research Funding; Kiowa]Kirin: Research Funding; Seattle Genetics: Consultancy, Research Funding; Spectrum: Research Funding; Amgen: Consultancy; Bristol]Myers Squibb,: Consultancy; Jannsen: Consultancy. Hamlin:Gilead, Spectrum, Seattle Genetics, Genentech: Consultancy; Spectrum, GSK, Jansen and Jansen/Pharmacyclics, Portola, Seattle Genetics: Research Funding. Matasar:Genentech: Consultancy; Spectrum: Consultancy. Miller:Foundation Medicine: Employment. Stephens:Foundation Medicine: Employment, Equity Ownership. He:Foundation Medicine: Employment. Younes:Novartis: Research Funding; J & J: Research Funding; Curis: Research Funding; Bayer; Bristol Meyer Squibb; Celgene; Incyte; Janssen R & D; Sanofi; Seattle Genetics; Takeda Millenium: Honoraria.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal