Abstract
Background.
The cAMP responsive element-binding (CREB) and Activating transcription factor (ATF) family of transcription factors regulates many cellular stress responses including proliferation, differentiation and survival, possibly through chromatin modification. CREB is a critical regulator of normal myelopoiesis and is over-expressed in AML, with knockdown inhibiting proliferation, suggesting a proto-oncogene role. CREB/ATF proteins have typically been studied individually , and in small series. Interactions with multiple signaling and functional pathways are suspected, but the actual relationship in primary AML samples is unknown. We therefore assessed the protein expression of two CREB/ATF family members; CREB binding protein 1 (CREB1) a leucine zipper transcription factor, active when phosphorylated on serine 133 (CREB1.pS133) and ATF 3 (ATF3), in a series of 511 newly diagnosed AML patients and compared expression to 228 simultaneously measured proteins
Methods. A reverse phase protein array (RPPA) using leukemia enriched cells from 511 AML patients. Both bone marrow (BM, n=387) and peripheral blood (PB, n=283) samples were used, with 140 cases having both. The RPPA was probed with 231 strictly validated antibodies, including antibodies against CREB1, phopsho CREB1 CREB1.pS133 and ATF3. Expression was compared to that of normal BM derived CD34+ cells. Interaction networks with the other 228 proteins were generated using glasso, supplemented by the literature of known interactions.
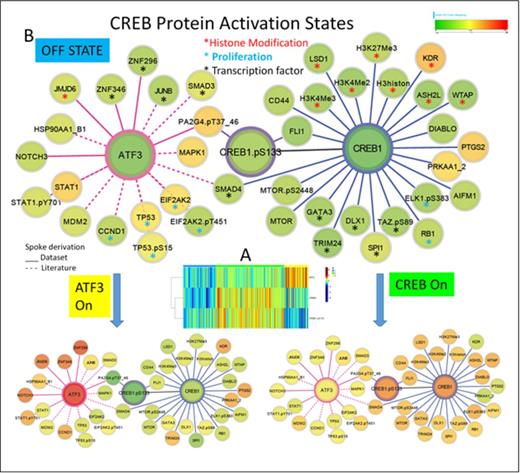
Results. A heatmap of CREB1,CREB1.pS133 and ATF expression was generated and k-means clustering performed (Figure A). Most cases of AML demonstrated high expression of CREB or ATF, but not both, although ATF levels were modest with high CREB1 expression, Using the Prototype Clusteringmethod an optimal division into three clusters C1) Pan Low expression C2) High CREB and C3) High ATF3 with 26%, 55% and 19% of patients in each respectively was selected. Consistent with the literature, expression of CREB1 & CREB1pS133 were strongly positively (Figure B) correlated with several histone modification proteins including Histone3, H3K4Me2, H3K4Me3, ASH2L, proliferation associated proteins RB1 and ELK1.pS383, and transcription factors including DLX1, Fli1, GATA3, Smad4, SPI1, TAZ.pS89 and Trim24. They were inversely correlated with histone demethylase KDR, protein kinase A and prostaglandin synthetase 2. ATF3 expression was positively correlated with histone modifier JMJD6, proliferation proteins EIF2AK2.pT451, CCND1 , and transcription factors JUNB, Smads 3 and 4, ZNF296, ZNF346. ATF levels were negatively correlated with Signal transduction via including STAT1, MAPK1, and PA2G4.pT37. While directionality cannot be inferred these proteins showed clear changes in expression of either ATF3 or CREB1/CREB1.pS133 between the pan off and the individual on states. WBC (14, 33, 30K respectively for C1,C2 and C3, p = 0.01) and %PB blasts (20, 34, 32%, p = 0.001) were significantly lower in C1, but most other clinical features including cytogenetics, and FLT3-ITD status did not differ between the clusters. Cluster membership was not associated with complete remission or primary resistance rates. Overall survival (OS) for all patients did not differ by cluster (p=0.45), but those with intermediate cytogenetics (IntCyto) and high CREB fared worse (median survival of 58 weeks vs. 66 and 87 for C3 and C1 (p=0.036) and this effect was more prominent in FLT3 mutant (p =0.05) than wildtype cases (p=0.34). Remission duration (RemDur) was similarly inferior in IntCyto C2 patients (median 39 vs. 76vs 89 weeks for C3 and C 1, p= 0.007)
Conclusions. Over expression of a CREB family member was very common in AML (74%) but was exclusive foreither ATF3 or CREB1, but not both. Overexpression was independent of clinical features, excluding higher WBC and %PBblast % with high CREB1. In those with IntCyto high CREB1 expression was adverse for OS and RemDur. High CREB1 or ATF expression correlated with histone modification, proliferation and transcription factor proteinexpression, but with different members of these protein functional classes. These results imply a central role for CREB/ATF family in deregulation of many pathways, but suggests that therapy directed towards interfering with CREB/ATF family must be selective to which side of the family is overexpressed to be effective.
Ravandi:Cellerant Therapeutics: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal